A Vernalization Response in a Winter Safflower (Carthamus tinctorius) Involves the Upregulation of Homologs of FT, FUL, and MAF
- PMID: 33859660
- PMCID: PMC8043130
- DOI: 10.3389/fpls.2021.639014
A Vernalization Response in a Winter Safflower (Carthamus tinctorius) Involves the Upregulation of Homologs of FT, FUL, and MAF
Abstract
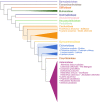
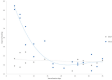
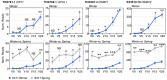
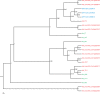
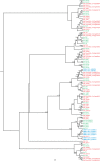
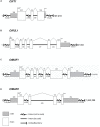
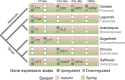
Safflower (Carthamus tinctorius) is a member of the Asteraceae family that is grown in temperate climates as an oil seed crop. Most commercially grown safflower varieties can be sown in late winter or early spring and flower rapidly in the absence of overwintering. There are winter-hardy safflower accessions that can be sown in autumn and survive over-wintering. Here, we show that a winter-hardy safflower possesses a vernalization response, whereby flowering is accelerated by exposing germinating seeds to prolonged cold. The impact of vernalization was quantitative, such that increasing the duration of cold treatment accelerated flowering to a greater extent, until the response was saturated after 2 weeks exposure to low-temperatures. To investigate the molecular-basis of the vernalization-response in safflower, transcriptome activity was compared and contrasted between vernalized versus non-vernalized plants, in both 'winter hardy' and 'spring' cultivars. These genome-wide expression analyses identified a small set of transcripts that are both differentially expressed following vernalization and that also have different expression levels in the spring versus winter safflowers. Four of these transcripts were quantitatively induced by vernalization in a winter hardy safflower but show high basal levels in spring safflower. Phylogenetic analyses confidently assigned that the nucleotide sequences of the four differentially expressed transcripts are related to FLOWERING LOCUS T (FT), FRUITFUL (FUL), and two genes within the MADS-like clade genes. Gene models were built for each of these sequences by assembling an improved safflower reference genome using PacBio-based long-read sequencing, covering 85% of the genome, with N50 at 594,000 bp in 3000 contigs. Possible evolutionary relationships between the vernalization response of safflower and those of other plants are discussed.
Keywords: PacBio; flowering time; genome; safflower; vernalization.
Copyright © 2021 Cullerne, Fjellheim, Spriggs, Eamens, Trevaskis and Wood.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Andrews S., Krueger F., Krueger C., Wingate S. (2012). FastQC. Babraham: Babraham Institute.
-
- Baloch D. M., Karow R. S., Marx E., Kling J. G., Witt M. D. (2003). Vernalization studies with Pacific Northwest wheat. Agron. J. 95 1201–1208. 10.2134/agronj2003.1201 - DOI
-
- Baskin J. M., Baskin C. C. (1989). Seed-germination ecophysiology of jeffersonia-diphylla, a perennial herb of mesic deciduous forests. Am. J. Bot. 76 1073–1080. 10.1002/j.1537-2197.1989.tb15088.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

