Targeting long noncoding RNA PMIF facilitates osteoprogenitor cells migrating to bone formation surface to promote bone formation during aging
- PMID: 33859765
- PMCID: PMC8039942
- DOI: 10.7150/thno.54477
Targeting long noncoding RNA PMIF facilitates osteoprogenitor cells migrating to bone formation surface to promote bone formation during aging
Abstract
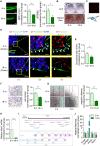
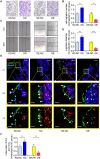
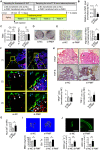
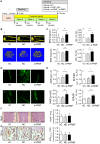
Rationale: The migration of mesenchymal osteoprogenitor cells (OPCs) to bone formation surface is the initial step of osteoblastogenesis before they undergo osteoblast differentiation and maturation for governing bone formation. However, whether the migration capacity of OPCs is compromised during aging and how it contributes to the aging-related bone formation reduction remain unexplored. In the present study, we identified a migration inhibitory factor (i.e., long noncoding RNA PMIF) and examined whether targeting lnc-PMIF could facilitate osteoprogenitor cells migrating to bone formation surface to promote bone formation during aging. Methods: Primary OPCs from young (6-momth-old) and aged (18-momth-old) C57BL/6 mice and stable lnc-PMIF knockdown/overexpression cell lines were used for in vitro and in vivo cell migration assay (i.e., wound healing assay, transwell assay and cell intratibial injection assay). RNA pulldown-MS/WB and RIP-qPCR were performed to identify the RNA binding proteins (RBPs) of lnc-PMIF. Truncations of lnc-PMIF and the identified RBP were engaged to determine the interaction motif between them by RNA pulldown-WB and EMSA. By cell-based therapy approach and by pharmacological approach, small interfering RNA (siRNA)-mediated lnc-PMIF knockdown were used in aged mice. The cell migration ability was evaluated by transwell assay and cell intratibial injection assay. The bone formation was evaluated by microCT analysis and bone morphometry analysis. Results: We reported that the decreased bone formation was accompanied by the reduced migration capacity of the bone marrow mesenchymal stem cells (BMSCs, the unique source of OPCs in bone marrow) in aged mice. We further identified that the long non-coding RNA PMIF (postulated migration inhibitory factor) (i.e., lnc-PMIF) was highly expressed in BMSCs from aged mice and responsible for the reduced migration capacity of aged OPCs to bone formation surface. Mechanistically, we found that lnc-PMIF could bind to human antigen R (HuR) for interrupting the HuR-β-actin mRNA interaction, therefore inhibit the expression of β-actin for suppressing the migration of aged OPCs. We also authenticated a functionally conserved human lncRNA ortholog of the murine lnc-PMIF. By cell-based therapy approach, we demonstrated that replenishing the aged BMSCs with small interfering RNA (siRNA)-mediated lnc-PMIF knockdown could promote bone formation in aged mice. By pharmacological approach, we showed that targeted delivery of lnc-PMIF siRNA approaching the OPCs around the bone formation surface could also promote bone formation in aged mice. Conclusion: Toward translational medicine, this study hints that targeting lnc-PMIF to facilitate aged OPCs migrating to bone formation surface could be a brand-new anabolic strategy for aging-related osteoporosis.
Keywords: aging; bone formation; cell migration; long non-coding RNA; osteoprogenitor cells.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


References
-
- Manolagas SC. Birth and death of bone cells: Basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–37. - PubMed
-
- Long FX. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. - PubMed
-
- Rosen CJ, Bouxsein ML. Mechanisms of disease: is osteoporosis the obesity of bone? Nat Clin Pract Rheumatol. 2006;2:35–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

