Precise detection of a murine germline mutation of the Notch3 gene associated with kyphosis and developmental disorders
- PMID: 33860007
- PMCID: PMC8043348
- DOI: 10.5455/javar.2021.h479
Precise detection of a murine germline mutation of the Notch3 gene associated with kyphosis and developmental disorders
Abstract
Objective: Humpback (hpbk) mice harbor a pathogenic mutation in the Notch3 gene and can serve as a beneficial animal model for investigating human myopathy, kyphosis, and developmental disorders, including lateral meningocele syndrome. Detection of the point mutation in hpbk mice is important for maintaining strains and scrutinizing genetic rescues, especially considering that homozygous mice are infertile and indistinguishable from their littermates at a young age. This study aimed for the development of a novel, precise, and time-saving genotyping method to identify the mutation in hpbk mice.
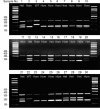
Materials and methods: In order to study the hpbk mouse line, we describe how we applied several tools, including quantitative polymerase chain reaction (qPCR), multiplex tetra-primer amplification-refractory mutation system (ARMS-PCR) and Sanger sequencing, toward the recognition of heterozygous and homozygous mice.
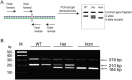
Results: The Notch3 mutation was clearly identified using qPCR and ARMS assays, but the latter was a more precise and cost-effective approach. The lengths of the ARMS-PCR amplicons are 210 bp and 164 bp for the wild-type and hpbk alleles, respectively. Moreover, the genotyping results for each mouse were corroborated by Sanger DNA sequencing.
Conclusion: Our newly developed PCR-based ARMS system affords a swift and precise way to genotype the hpbk mice. ARMS-PCR does not rely on any advanced equipment and is useful as a genotyping method for other model organisms that harbor a pathogenic variant.
Keywords: ARMS; Lateral meningocele syndrome; Notch3 mutation; genotyping; skeletal disease.
Copyright: © Journal of Advanced Veterinary and Animal Research.
Conflict of interest statement
The authors declare no financial conflict of interests.
Figures



Similar articles
-
A Tetra-Primer Amplification Refractory System Technique for the Cost-Effective and Novel Genotyping of Eight Single-Nucleotide Polymorphisms of the Catechol-O-Methyltransferase Gene.Genet Test Mol Biomarkers. 2016 Aug;20(8):465-70. doi: 10.1089/gtmb.2015.0304. Epub 2016 May 26. Genet Test Mol Biomarkers. 2016. PMID: 27228319
-
Rapid and efficient identification of the mouse leptin receptor mutation (C57BL/KsJ-db/db) by tetra-primer amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) analysis.Lab Anim Res. 2016 Mar;32(1):70-3. doi: 10.5625/lar.2016.32.1.70. Epub 2016 Mar 24. Lab Anim Res. 2016. PMID: 27051445 Free PMC article.
-
Tetra-Primer Amplification-Refractory Mutation System (ARMS)-PCR for Genotyping Mouse Leptin Gene Mutation.Animals (Basel). 2022 Oct 5;12(19):2680. doi: 10.3390/ani12192680. Animals (Basel). 2022. PMID: 36230421 Free PMC article.
-
Conventional polymerase chain reaction and amplification refractory mutation system-multi-gene/ multi-primer PCR in the diagnosis of female genital tuberculosis.Arch Microbiol. 2019 Apr;201(3):267-281. doi: 10.1007/s00203-019-01631-1. Epub 2019 Feb 20. Arch Microbiol. 2019. PMID: 30788519 Review.
-
COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing.Biochem Soc Trans. 2009 Apr;37(Pt 2):427-32. doi: 10.1042/BST0370427. Biochem Soc Trans. 2009. PMID: 19290875 Review.
Cited by
-
Congenital kyphoscoliosis: Analysis of vertebral abnormalities using model animals (Review).Exp Ther Med. 2024 Sep 4;28(5):416. doi: 10.3892/etm.2024.12705. eCollection 2024 Nov. Exp Ther Med. 2024. PMID: 39301254 Free PMC article. Review.
References
-
- Mašek J, Andersson ER. The developmental biology of genetic Notch disorders. Dev Suppl. 2017;144(10):1743–63. https://doi.org/10.1242/dev.148007. - PubMed
-
- Fairfield H, Gilbert GJ, Barter M, Corrigan RR, Curtain M, Ding Y, et al. Mutation discovery in mice by whole exome sequencing. Genome Biol. 2011;12(9):R86. https://doi.org/10.1186/gb-2011-12-9-r86. - PMC - PubMed
-
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. https://doi.org/10.1016/j.cell.2009.03.045. - PMC - PubMed
-
- Tao J, Chen S, Lee B. Alteration of Notch signaling in skeletal development and disease. Ann N Y Acad Sci. 2010;1192:257–68. https://doi.org/10.1111/j.1749-6632.2009.05307.x. - PMC - PubMed
-
- Gripp KW, Robbins KM, Sobreira NL, Witmer PD, Bird LM, Avela K, et al. Truncating mutations in the last exon of NOTCH3 cause lateral meningocele syndrome. Am J Med Genet Part A. 2015;167A(2):271–81. https://doi.org/10.1002/ajmg.a.36863. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
