Digital next-generation sequencing of cell-free DNA for pancreatic cancer
- PMID: 33860102
- PMCID: PMC8035455
- DOI: 10.1002/jgh3.12530
Digital next-generation sequencing of cell-free DNA for pancreatic cancer
Abstract
Background and aim: The clinical applicability of digital next-generation sequencing (dNGS), which eliminates polymerase chain reaction (PCR) and sequencing error-derived noise by using molecular barcodes (MBs), has not been fully evaluated. We evaluated the utility of dNGS of cell-free DNA (cfDNA) in liquid biopsies obtained from patients with pancreatic cancer.
Methods: Fifty-eight patients with pancreatic cancer undergoing endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) were included. Samples were subjected to sequencing of 50 cancer-related genes using next-generation sequencing (NGS). The results were used as reference gene alterations. NGS of cfDNA from plasma was performed for patients with a mutant allele frequency (MAF) >1% and an absolute mutant number > 10 copies/plasma mL in KRAS or GNAS by digital PCR. Sequence readings with and without MBs were compared with reference to EUS-FNA-derived gene alterations.
Results: The concordance rate between dNGS of cfDNA and EUS-FNA-derived gene alterations was higher with than without MBs (p = 0.039), and MAF cut-off values in dNGS could be decreased to 0.2%. dNGS using MBs eliminated PCR and sequencing error by 74% and 68% for TP53 and all genes, respectively. Overall, dNGS detected mutations in KRAS (45%) and TP53 (26%) and copy number alterations in CCND2, CCND3, CDK4, FGFR1, and MYC, which are targets of molecular-targeted drugs.
Conclusions: dNGS of cfDNA using MBs is useful for accurate detection of gene alterations even with low levels of MAFs. These results may be used to inform the development of diagnostics and therapeutics that can improve the prognosis of pancreatic cancer.
Keywords: cell‐free DNA; liquid biopsy; next‐generation sequencing; pancreatic ductal carcinoma.
© 2021 The Authors. JGH Open published by Journal of Gastroenterology and Hepatology Foundation and John Wiley & Sons Australia, Ltd.
Figures



 , With MB;
, With MB;  , without MB. (b) MAF plots of detected mutations in cfDNA by next‐generation sequencing (NGS) analyses with and without MB. Matches and mismatches with mutations detected in tissue samples are shown in blue and orange dots, respectively. Cut‐off values for matched mutations (i.e. same mutations in cfDNA and tissue samples) were determined by ROC curve analysis to be 0.2% and 0.5% by dNGS and ordinary NGS, respectively. The sensitivity and specificity for matched mutations were 83.3% and 93.3% by dNGS, respectively, and 73.9% and 92.9% by ordinary NGS, respectively.
, without MB. (b) MAF plots of detected mutations in cfDNA by next‐generation sequencing (NGS) analyses with and without MB. Matches and mismatches with mutations detected in tissue samples are shown in blue and orange dots, respectively. Cut‐off values for matched mutations (i.e. same mutations in cfDNA and tissue samples) were determined by ROC curve analysis to be 0.2% and 0.5% by dNGS and ordinary NGS, respectively. The sensitivity and specificity for matched mutations were 83.3% and 93.3% by dNGS, respectively, and 73.9% and 92.9% by ordinary NGS, respectively.  , Match;
, Match;  , mismatch. (c) The proportion of concomitant cfDNA with tissue mutations in KRAS, TP53, and all assayed genes. The number of mutations detected in tissue samples are shown in the left in each graph, and cfDNA mutations matching with those identified in tissue samples are shown in bar plots. Tissue‐matched and ‐mismatched cfDNA mutations are shown as blue and yellow bars, respectively, whereas tissue‐matched mutations with a lower MAF than the cut‐off value are shown as light green bars.
, mismatch. (c) The proportion of concomitant cfDNA with tissue mutations in KRAS, TP53, and all assayed genes. The number of mutations detected in tissue samples are shown in the left in each graph, and cfDNA mutations matching with those identified in tissue samples are shown in bar plots. Tissue‐matched and ‐mismatched cfDNA mutations are shown as blue and yellow bars, respectively, whereas tissue‐matched mutations with a lower MAF than the cut‐off value are shown as light green bars.  , Mismatch;
, Mismatch;  , match < cutoff;
, match < cutoff;  , match or tissue. MAF, mutant allele frequency; MB, dNGS analysis with molecular barcodes; cfDNA, cell‐free DNA.
, match or tissue. MAF, mutant allele frequency; MB, dNGS analysis with molecular barcodes; cfDNA, cell‐free DNA.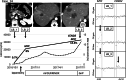
References
-
- Vogelzang NJ, Benowitz SI, Adams S et al. Clinical cancer advances 2011: annual report on progress against cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 2012; 30: 88–109. - PubMed
-
- Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: treating cancer with specificity. Eur. J. Pharmacol. 2018; 834: 188–96. - PubMed
-
- Shin HJ, Lahoti S, Sneige N. Endoscopic ultrasound‐guided fine‐needle aspiration in 179 cases: the M. D. Anderson Cancer Center experience. Cancer. 2002; 96: 174–80. - PubMed
-
- Forshew T, Murtaza M, Parkinson C et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012; 4: 136ra68. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

