Establishment of a Cell Culture Model Permissive for Infection by Hepatitis B and C Viruses
- PMID: 33860122
- PMCID: PMC8034569
- DOI: 10.1002/hep4.1653
Establishment of a Cell Culture Model Permissive for Infection by Hepatitis B and C Viruses
Abstract
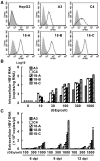
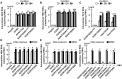
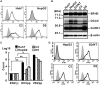
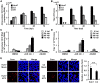
Compared with each monoinfection, coinfection with hepatitis B virus (HBV) and hepatitis C virus (HCV) is well known to increase the risks of developing liver cirrhosis and hepatocellular carcinoma. However, the mechanism by which HBV/HCV coinfection is established in hepatocytes is not well understood. Common cell culture models for coinfection are required to examine viral propagation. In this study, we aimed to establish a cell line permissive for both HBV and HCV infection. We first prepared a HepG2 cell line expressing sodium taurocholate cotransporting polypeptide, an HBV receptor, and then selected a cell line highly permissive for HBV infection, G2/NT18-B. After transduction with a lentivirus-encoding microRNA-122, the cell line harboring the highest level of replicon RNA was selected and then treated with anti-HCV compounds to eliminate the replicon RNA. The resulting cured cell line was transduced with a plasmid-encoding CD81. The cell line permissive for HCV infection was cloned and then designated the G2BC-C2 cell line, which exhibited permissiveness for HBV and HCV propagation. JAK inhibitor I potentiated the HCV superinfection of HBV-infected cells, and fluorescence-activated cell-sorting analysis indicated that HBV/HCV double-positive cells accounted for approximately 30% of the coinfected cells. Among several host genes tested, cyclooxygenase-2 showed synergistic induction by coinfection compared with each monoinfection. Conclusion: These data indicate that our in vitro HBV/HCV coinfection system provides an easy-to-use platform for the study of host and viral responses against coinfection and the development of antiviral agents targeting HBV and HCV.
© 2020 The Authors. Hepatology Communications published by Wiley Periodicals LLC on behalf of the American Association for the Study of Liver Diseases.
Figures



Similar articles
-
A novel cell culture model reveals the viral interference during hepatitis B and C virus coinfection.Antiviral Res. 2021 May;189:105061. doi: 10.1016/j.antiviral.2021.105061. Epub 2021 Mar 9. Antiviral Res. 2021. PMID: 33705864
-
Robust Human and Murine Hepatocyte Culture Models of Hepatitis B Virus Infection and Replication.J Virol. 2018 Nov 12;92(23):e01255-18. doi: 10.1128/JVI.01255-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30232184 Free PMC article.
-
HBV Bypasses the Innate Immune Response and Does Not Protect HCV From Antiviral Activity of Interferon.Gastroenterology. 2018 May;154(6):1791-1804.e22. doi: 10.1053/j.gastro.2018.01.044. Epub 2018 Feb 1. Gastroenterology. 2018. PMID: 29410097
-
Hepatitis B virus (HBV) reactivation-The potential role of direct-acting agents for hepatitis C virus (HCV).Rev Med Virol. 2018 Jul;28(4):e1984. doi: 10.1002/rmv.1984. Epub 2018 May 15. Rev Med Virol. 2018. PMID: 29761585 Free PMC article. Review.
-
Epidemiology, natural history, and treatment of hepatitis B virus and hepatitis C virus coinfection.Minerva Gastroenterol Dietol. 2006 Jun;52(2):145-55. Minerva Gastroenterol Dietol. 2006. PMID: 16557186 Review.
Cited by
-
Relevance of HBx for Hepatitis B Virus-Associated Pathogenesis.Int J Mol Sci. 2023 Mar 4;24(5):4964. doi: 10.3390/ijms24054964. Int J Mol Sci. 2023. PMID: 36902395 Free PMC article. Review.
-
Forthcoming Developments in Models to Study the Hepatitis B Virus Replication Cycle, Pathogenesis, and Pharmacological Advancements.ACS Omega. 2023 Apr 14;8(16):14273-14289. doi: 10.1021/acsomega.2c07154. eCollection 2023 Apr 25. ACS Omega. 2023. PMID: 37125123 Free PMC article. Review.
-
Neutralization of hepatitis B virus with vaccine-escape mutations by hepatitis B vaccine with large-HBs antigen.Nat Commun. 2022 Sep 5;13(1):5207. doi: 10.1038/s41467-022-32910-z. Nat Commun. 2022. PMID: 36064848 Free PMC article.
-
Establishment of a stable SARS-CoV-2 replicon system for application in high-throughput screening.Antiviral Res. 2022 Mar;199:105268. doi: 10.1016/j.antiviral.2022.105268. Epub 2022 Mar 7. Antiviral Res. 2022. PMID: 35271914 Free PMC article.
-
Production and Purification of Cell Culture-generated Hepatitis B Virus by Transient Transfection and Density Gradient.Bio Protoc. 2023 Jul 20;13(14):e4779. doi: 10.21769/BioProtoc.4779. eCollection 2023 Jul 20. Bio Protoc. 2023. PMID: 37497458 Free PMC article.
References
-
- Ward JW, Hinman AR. What is needed to eliminate hepatitis B virus and hepatitis C virus as global health threats. Gastroenterology 2019;156:297‐310. - PubMed
-
- Abdelaal R, Yanny B, El Kabany M. HBV/HCV coinfection in the era of HCV‐DAAs. Clin Liver Dis 2019;23:463‐472. - PubMed
-
- Kawagishi N, Suda G, Onozawa M, Kimura M, Maehara O, Ohara M, et al. Comparing the risk of hepatitis B virus reactivation between direct‐acting antiviral therapies and interferon‐based therapies for hepatitis C. J Viral Hepat 2017;24:1098‐1106. - PubMed
-
- Sagnelli E, Coppola N, Messina V, Di Caprio D, Marrocco C, Marotta A, et al. HBV superinfection in hepatitis C virus chronic carriers, viral interaction, and clinical course. Hepatology 2002;36:1285‐1291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

