Mass spectrometry-based proteomic platforms for better understanding of SARS-CoV-2 induced pathogenesis and potential diagnostic approaches
- PMID: 33860983
- PMCID: PMC8250252
- DOI: 10.1002/pmic.202000279
Mass spectrometry-based proteomic platforms for better understanding of SARS-CoV-2 induced pathogenesis and potential diagnostic approaches
Abstract
While protein-protein interaction is the first step of the SARS-CoV-2 infection, recent comparative proteomic profiling enabled the identification of over 11,000 protein dynamics, thus providing a comprehensive reflection of the molecular mechanisms underlying the cellular system in response to viral infection. Here we summarize and rationalize the results obtained by various mass spectrometry (MS)-based proteomic approaches applied to the functional characterization of proteins and pathways associated with SARS-CoV-2-mediated infections in humans. Comparative analysis of cell-lines versus tissue samples indicates that our knowledge in proteome profile alternation in response to SARS-CoV-2 infection is still incomplete and the tissue-specific response to SARS-CoV-2 infection can probably not be recapitulated efficiently by in vitro experiments. However, regardless of the viral infection period, sample types, and experimental strategies, a thorough cross-comparison of the recently published proteome, phosphoproteome, and interactome datasets led to the identification of a common set of proteins and kinases associated with PI3K-Akt, EGFR, MAPK, Rap1, and AMPK signaling pathways. Ephrin receptor A2 (EPHA2) was identified by 11 studies including all proteomic platforms, suggesting it as a potential future target for SARS-CoV-2 infection mechanisms and the development of new therapeutic strategies. We further discuss the potentials of future proteomics strategies for identifying prognostic SARS-CoV-2 responsive age-, gender-dependent, tissue-specific protein targets.
Keywords: COVID-19; biomarkers; comparative proteomics; kinase-substrate signaling; post-translational modifications; targeted proteomics; top-down proteomics.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
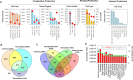
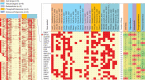
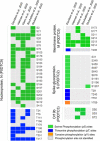


References
-
- Wadman, M. , Couzin‐Frankel, J. , Kaiser, J. , & Matacic, C. (2020). A Rampage Through the Body. Science, 368, 356–360. - PubMed
-
- Appelberg, S. , Gupta, S. , Akusjärvi, S. S. , Ambikan, A. T. , Mikaeloff, F. , Saccon, E. , Végvári, Á. , Benfeitas, R. , Sperk, M. , Ståhlberg, M. , Krishnan, S. , Singh, K. , Penninger, J. M. , Mirazimi, A. , & Neogi, U. (2020). Dysregulation in Akt/mTOR/HIF‐1 signaling identified by proteo‐transcriptomics of SARS‐CoV‐2 infected cells. Emerg Microbes Infect, 9, 1748–1760. - PMC - PubMed
-
- Grenga, L. , Gallais, F. , Pible, O. , Gaillard, J. C. , Gouveia, D. , Batina, H. , Bazaline, N. , Ruat, S. , Culotta, K. , Miotello, G. , Debroas, S. , Roncato, M. A. , Steinmetz, G. , Foissard, C. , Desplan, A. , Alpha‐Bazin, B. , Almunia, C. , Gas, F. , Bellanger, L. , & Armengaud, J. (2020). Shotgun proteomics analysis of SARS‐CoV‐2‐infected cells and how it can optimize whole viral particle antigen production for vaccines. Emerg Microbes Infect, 9, 1712–1721. - PMC - PubMed
-
- Zecha, J. , Lee, C. Y. , Bayer, F. P. , Meng, C. , Grass, V. , Zerweck, J. , Schnatbaum, K. , Michler, T. , Pichlmair, A. , Ludwig, C. , & Kuster, B. (2020). Data, reagents, assays and merits of proteomics for SARS‐CoV‐2 research and testing. Molecular & Cellular Proteomics, 19, 1503–1522. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

