FRugally Optimized DNA Octomer (FRODO) qPCR Measurement of HIV and SIV in Human and Nonhuman Primate Samples
- PMID: 33861500
- PMCID: PMC8054980
- DOI: 10.1002/cpz1.93
FRugally Optimized DNA Octomer (FRODO) qPCR Measurement of HIV and SIV in Human and Nonhuman Primate Samples
Abstract
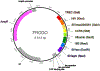
Quantitative polymerase chain reactions (qPCRs) are commonly employed to enumerate genes of interest among particular biological samples. Insertion of PCR amplicons into plasmid DNA is a mainstay for creation of known quantities of target sequences to standardize qPCRs. Typically, one amplicon is inserted into one plasmid construct, and the plasmid is then amplified, purified, serially diluted, and quantified to be used to enumerate target sequences in unknown samples. As qPCR is often used to detect multiple amplicons simultaneously, individual qPCR standards are often desired to normalize one to another. Here we report a single plasmid containing eight amplicons, which can be used to quantify several different strains of simian immunodeficiency virus and human immunodeficiency virus, cell number equivalents for humans and nonhuman primates, T cell receptor excision circles, and bacterial 16S DNA. This FRugally Optimized DNA Octomer (FRODO) plasmid was created and standardized to quantify all eight PCR amplicons. © 2021 US Government. Basic Protocol 1: Total genomic DNA extraction from primary cells Basic Protocol 2: Quantitative PCR for viral, bacterial, and cell number equivalents Support Protocol: Purification, quantification, and storage of FRODO standard plasmid DNA.
Keywords: HIV; SIV; nonhuman primate; qPCR.
Published 2021. This article is a U.S. Government work and is in the public domain in the USA.
Figures


References
-
- Brenchley JM, Vinton C, Tabb B, Hao XP, Connick E, Paiardini M, … Estes JD (2012). Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood, 120(20), 4172–4181. doi:10.1182/blood-2012-06-437608 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

