Genome-wide analysis of lncRNA stability in human
- PMID: 33861746
- PMCID: PMC8081339
- DOI: 10.1371/journal.pcbi.1008918
Genome-wide analysis of lncRNA stability in human
Abstract
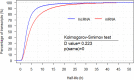
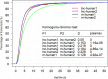
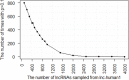
Transcript stability is associated with many biological processes, and the factors affecting mRNA stability have been extensively studied. However, little is known about the features related to human long noncoding RNA (lncRNA) stability. By inhibiting transcription and collecting samples in 10 time points, genome-wide RNA-seq studies was performed in human lung adenocarcinoma cells (A549) and RNA half-life datasets were constructed. The following observations were obtained. First, the half-life distributions of both lncRNAs and messanger RNAs (mRNAs) with one exon (lnc-human1 and m-human1) were significantly different from those of both lncRNAs and mRNAs with more than one exon (lnc-human2 and m-human2). Furthermore, some factors such as full-length transcript secondary structures played a contrary role in lnc-human1 and m-human2. Second, through the half-life comparisons of nucleus- and cytoplasm-specific and common lncRNAs and mRNAs, lncRNAs (mRNAs) in the nucleus were found to be less stable than those in the cytoplasm, which was derived from transcripts themselves rather than cellular location. Third, kmers-based protein-RNA or RNA-RNA interactions promoted lncRNA stability from lnc-human1 and decreased mRNA stability from m-human2 with high probability. Finally, through applying deep learning-based regression, a non-linear relationship was found to exist between the half-lives of lncRNAs (mRNAs) and related factors. The present study established lncRNA and mRNA half-life regulation networks in the A549 cell line and shed new light on the degradation behaviors of both lncRNAs and mRNAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. P Natl Acad Sci USA. 2002;99(15):9697–702. 10.1073/pnas.112318199 PubMed PMID: WOS:000177042400020. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

