The position of single-base deletions in the VNTR sequence of the carboxyl ester lipase (CEL) gene determines proteotoxicity
- PMID: 33862081
- PMCID: PMC8692231
- DOI: 10.1016/j.jbc.2021.100661
The position of single-base deletions in the VNTR sequence of the carboxyl ester lipase (CEL) gene determines proteotoxicity
Abstract
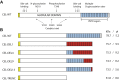
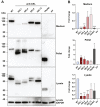
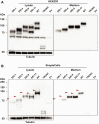
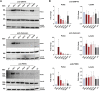
Variable number of tandem repeat (VNTR) sequences in the genome can have functional consequences that contribute to human disease. This is the case for the CEL gene, which is specifically expressed in pancreatic acinar cells and encodes the digestive enzyme carboxyl ester lipase. Rare single-base deletions (DELs) within the first (DEL1) or fourth (DEL4) VNTR segment of CEL cause maturity-onset diabetes of the young, type 8 (MODY8), an inherited disorder characterized by exocrine pancreatic dysfunction and diabetes. Studies on the DEL1 variant have suggested that MODY8 is initiated by CEL protein misfolding and aggregation. However, it is unclear how the position of single-base deletions within the CEL VNTR affects pathogenic properties of the protein. Here, we investigated four naturally occurring CEL variants, arising from single-base deletions in different VNTR segments (DEL1, DEL4, DEL9, and DEL13). When the four variants were expressed in human embryonic kidney 293 cells, only DEL1 and DEL4 led to significantly reduced secretion, increased intracellular aggregation, and increased endoplasmic reticulum stress compared with normal CEL protein. The level of O-glycosylation was affected in all DEL variants. Moreover, all variants had enzymatic activity comparable with that of normal CEL. We conclude that the longest aberrant protein tails, resulting from single-base deletions in the proximal VNTR segments, have highest pathogenic potential, explaining why DEL1 and DEL4 but not DEL9 and DEL13 have been observed in patients with MODY8. These findings further support the view that CEL mutations cause pancreatic disease through protein misfolding and proteotoxicity, leading to endoplasmic reticulum stress and activation of the unfolded protein response.
Keywords: CEL; MODY8; O-glycosylation; endoplasmic reticulum stress; protein misfolding; single-base deletions; unfolded protein response.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Brookes K.J. The VNTR in complex disorders: The forgotten polymorphisms? A functional way forward? Genomics. 2013;101:273–281. - PubMed
-
- Jeffreys A.J., Wilson V., Thein S.L. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985;316:76–79. - PubMed
-
- Simpson J., Vetuz G., Wilson M., Brookes K.J., Kent L. The DRD4 receptor Exon 3 VNTR and 5' SNP variants and mRNA expression in human post-mortem brain tissue. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153:1228–1233. - PubMed
-
- Johansson B.B., Fjeld K., El Jellas K., Gravdal A., Dalva M., Tjora E., Ræder H., Kulkarni R.N., Johansson S., Njølstad P.R., Molven A. The role of the carboxyl ester lipase (CEL) gene in pancreatic disease. Pancreatology. 2018;18:12–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

