Longitudinal Single-Cell Transcriptomics Reveals a Role for Serpina3n-Mediated Resolution of Inflammation in a Mouse Colitis Model
- PMID: 33862275
- PMCID: PMC8258998
- DOI: 10.1016/j.jcmgh.2021.04.004
Longitudinal Single-Cell Transcriptomics Reveals a Role for Serpina3n-Mediated Resolution of Inflammation in a Mouse Colitis Model
Abstract
Background & aims: Proper resolution of inflammation is essential to maintaining homeostasis, which is important as a dysregulated inflammatory response has adverse consequences, even being regarded as a hallmark of cancer. However, our picture of dynamic changes during inflammation remains far from comprehensive.
Methods: Here we used single-cell transcriptomics to elucidate changes in distinct cell types and their interactions in a mouse model of chemically induced colitis.
Results: Our analysis highlights the stromal cell population of the colon functions as a hub with dynamically changing roles over time. Importantly, we found that Serpina3n, a serine protease inhibitor, is specifically expressed in stromal cell clusters as inflammation resolves, interacting with a potential target, elastase. Indeed, genetic ablation of the Serpina3n gene delays resolution of induced inflammation. Furthermore, systemic Serpina3n administration promoted the resolution of inflammation, ameliorating colitis symptoms.
Conclusions: This study provides a comprehensive, single-cell understanding of cell-cell interactions during colorectal inflammation and reveals a potential therapeutic target that leverages inflammation resolution.
Keywords: Colitis; Serpina3n; Single Cell RNA-Sequencing; Stromal Cell.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

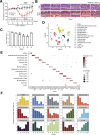




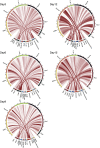

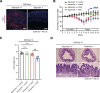


Comment in
-
A Cellular "Hub" Function to Resolve Colitis.Cell Mol Gastroenterol Hepatol. 2021;12(2):789-790. doi: 10.1016/j.jcmgh.2021.04.008. Epub 2021 May 8. Cell Mol Gastroenterol Hepatol. 2021. PMID: 33971162 Free PMC article. No abstract available.
References
-
- de Souza H.S.P., Fiocchi C., Iliopoulos D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol. 2017;14:739–749. - PubMed
-
- Henderson P., van Limbergen J.E., Schwarze J., Wilson D.C. Function of the intestinal epithelium and its dysregulation in inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:382–395. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

