Cross-oncopanel study reveals high sensitivity and accuracy with overall analytical performance depending on genomic regions
- PMID: 33863344
- PMCID: PMC8051090
- DOI: 10.1186/s13059-021-02315-0
Cross-oncopanel study reveals high sensitivity and accuracy with overall analytical performance depending on genomic regions
Abstract
Background: Targeted sequencing using oncopanels requires comprehensive assessments of accuracy and detection sensitivity to ensure analytical validity. By employing reference materials characterized by the U.S. Food and Drug Administration-led SEquence Quality Control project phase2 (SEQC2) effort, we perform a cross-platform multi-lab evaluation of eight Pan-Cancer panels to assess best practices for oncopanel sequencing.
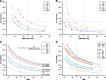
Results: All panels demonstrate high sensitivity across targeted high-confidence coding regions and variant types for the variants previously verified to have variant allele frequency (VAF) in the 5-20% range. Sensitivity is reduced by utilizing VAF thresholds due to inherent variability in VAF measurements. Enforcing a VAF threshold for reporting has a positive impact on reducing false positive calls. Importantly, the false positive rate is found to be significantly higher outside the high-confidence coding regions, resulting in lower reproducibility. Thus, region restriction and VAF thresholds lead to low relative technical variability in estimating promising biomarkers and tumor mutational burden.
Conclusion: This comprehensive study provides actionable guidelines for oncopanel sequencing and clear evidence that supports a simplified approach to assess the analytical performance of oncopanels. It will facilitate the rapid implementation, validation, and quality control of oncopanels in clinical use.
Keywords: Analytical performance; Molecular diagnostics; Oncopanel sequencing; Precision medicine; Reproducibility; Target enrichment.
Conflict of interest statement
The authors declare the following potential competing financial interests:
Natalia Novoradovskaya, Katherine Wilkins, Anne Lucas, Scott Happe, and Carlos Pabon are all employees of Agilent Technologies, Inc. Agilent sample B DNA reference sample is a current product and sample A DNA and sample C DNA are potential products of Agilent Technologies, Inc.
Figures




References
-
- Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8(7):823–59. 10.1097/JTO.0b013e318290868f. - PubMed
-
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Hersey P, Kefford R, Lawrence D, Puzanov I, Lewis KD, Amaravadi RK, Chmielowski B, Lawrence HJ, Shyr Y, Ye F, Li J, Nolop KB, Lee RJ, Joe AK, Ribas A. Survival in BRAF V600–mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. - DOI - PMC - PubMed
-
- Cordova-Delgado M, Pinto MP, Retamal IN, Muñoz-Medel M, Bravo ML, Fernández MF, Cisternas B, Mondaca S, Sanchez C, Galindo H, Nervi B, Ibáñez C, Acevedo F, Madrid J, Peña J, Koch E, Maturana MJ, Romero D, de la Jara N, Torres J, Espinoza M, Balmaceda C, Liao Y, Li Z, Freire M, Gárate-Calderón V, Cáceres J, Sepúlveda-Hermosilla G, Lizana R, Ramos L, Artigas R, Norero E, Crovari F, Armisén R, Corvalán AH, Owen GI, Garrido M. High proportion of potential candidates for immunotherapy in a Chilean cohort of gastric cancer patients: results of the FORCE1 study. Cancers. 2019;11(9):1275. doi: 10.3390/cancers11091275. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

