Experimental parameters defining ultra-low biomass bioaerosol analysis
- PMID: 33863892
- PMCID: PMC8052325
- DOI: 10.1038/s41522-021-00209-4
Experimental parameters defining ultra-low biomass bioaerosol analysis
Abstract
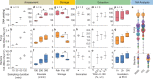
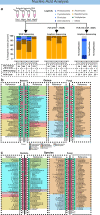
Investigation of the microbial ecology of terrestrial, aquatic and atmospheric ecosystems requires specific sampling and analytical technologies, owing to vastly different biomass densities typically encountered. In particular, the ultra-low biomass nature of air presents an inherent analytical challenge that is confounded by temporal fluctuations in community structure. Our ultra-low biomass pipeline advances the field of bioaerosol research by significantly reducing sampling times from days/weeks/months to minutes/hours, while maintaining the ability to perform species-level identification through direct metagenomic sequencing. The study further addresses all experimental factors contributing to analysis outcome, such as amassment, storage and extraction, as well as factors that impact on nucleic acid analysis. Quantity and quality of nucleic acid extracts from each optimisation step are evaluated using fluorometry, qPCR and sequencing. Both metagenomics and marker gene amplification-based (16S and ITS) sequencing are assessed with regard to their taxonomic resolution and inter-comparability. The pipeline is robust across a wide range of climatic settings, ranging from arctic to desert to tropical environments. Ultimately, the pipeline can be adapted to environmental settings, such as dust and surfaces, which also require ultra-low biomass analytics.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Darwin, C. The Voyage of the Beagle (Cosimo Inc., 2008).
-
- Von Humboldt, A. & Aimé B. Personal Narrative of Travels to the Equinoctial Regions of America: During the Years 1799-1804 (Cosimo Inc., 2013).
-
- Silvia, C. M. & Stal. J. L. The Marine Microbiome (Springer International, 2016).
-
- Burrows SM, Elbert W, Lawrence MG. Bacteria in the global atmosphere. Atmos. Chem. Phys. 2009;9:9263–9280. doi: 10.5194/acp-9-9263-2009. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

