Modeling muscle regeneration in RNA toxicity mice
- PMID: 33864373
- PMCID: PMC8188403
- DOI: 10.1093/hmg/ddab108
Modeling muscle regeneration in RNA toxicity mice
Abstract
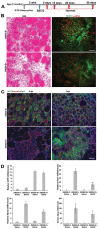
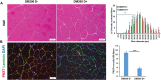
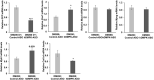
RNA toxicity underlies the pathogenesis of disorders such as myotonic dystrophy type 1 (DM1). Muscular dystrophy is a key element of the pathology of DM1. The means by which RNA toxicity causes muscular dystrophy in DM1 is unclear. Here, we have used the DM200 mouse model of RNA toxicity due to the expression of a mutant DMPK 3'UTR mRNA to model the effects of RNA toxicity on muscle regeneration. Using a BaCl2-induced damage model, we find that RNA toxicity leads to decreased expression of PAX7, and decreased numbers of satellite cells, the stem cells of adult skeletal muscle (also known as MuSCs). This is associated with a delay in regenerative response, a lack of muscle fiber maturation and an inability to maintain a normal number of satellite cells. Repeated muscle damage also elicited key aspects of muscular dystrophy, including fat droplet deposition and increased fibrosis, and the results represent one of the first times to model these classic markers of dystrophic changes in the skeletal muscles of a mouse model of RNA toxicity. Using a ligand-conjugated antisense (LICA) oligonucleotide ASO targeting DMPK sequences for the first time in a mouse model of RNA toxicity in DM1, we find that treatment with IONIS 877864, which targets the DMPK 3'UTR mRNA, is efficacious in correcting the defects in regenerative response and the reductions in satellite cell numbers caused by RNA toxicity. These results demonstrate the possibilities for therapeutic interventions to mitigate the muscular dystrophy associated with RNA toxicity in DM1.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures








References
-
- Day, J.W. and Ranum, L.P. (2005) RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord., 15, 5–16. - PubMed
-
- Mahadevan, M., Tsilfidis, C., Sabourin, L., Shutler, G., Amemiya, C., Jansen, G., Neville, C., Narang, M., Barcelo, J., O'Hoy, K. et al. (1992) Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science, 255, 1253–1255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

