Computational analysis of deleterious single nucleotide polymorphisms in catechol O-Methyltransferase conferring risk to post-traumatic stress disorder
- PMID: 33865170
- PMCID: PMC8969201
- DOI: 10.1016/j.jpsychires.2021.03.048
Computational analysis of deleterious single nucleotide polymorphisms in catechol O-Methyltransferase conferring risk to post-traumatic stress disorder
Abstract
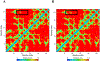
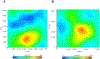
Post-traumatic stress disorder (PTSD) is one of the prevalent neurological disorder which is drawing increased attention over the past few decades. Major risk factors for PTSD can be categorized into environmental and genetic factors. Among the genetic risk factors, polymorphisms in the catechol-O-methyltransferase (COMT) gene is known to be associated with the risk for PTSD. In the present study, we analysed the impact of deleterious single nucleotide polymorphisms (SNPs) in the COMT gene conferring risk to PTSD using computational based approaches followed by molecular dynamic simulations. The data on COMT gene associated with PTSD were collected from several databases including Online Mendelian Inheritance in Man (OMIM) search. Datasets related to SNP were downloaded from the dbSNP database. To study the structural and dynamic effects of COMT wild type and mutant forms, we performed molecular dynamics simulations (MD simulations) at a time scale of 300 ns. Results from screening the SNPs using the computational tools SIFT and Polyphen-2 demonstrated that the SNP rs4680 (V158M) in COMT has a deleterious effect with phenotype in PTSD. Results from the MD simulations showed that there is some major fluctuations in the structural features including root mean square deviation (RMSD), radius of gyration (Rg), root mean square fluctuation (RMSF) and secondary structural elements including α-helices, sheets and turns between wild-type (WT) and mutant forms of COMT protein. In conclusion, our study provides novel insights into the deleterious effects and impact of V158M mutation on COMT protein structure which plays a key role in PTSD.
Keywords: Catechol-O-Methyltransferase (COMT); Molecular dynamic simulations (MD simulations); Molecular modelling; Mutation; Single nucleotide polymorphism (SNP).
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest
The authors declare no conflicts of interest.
Figures


Similar articles
-
COMT Val158Met polymorphism is associated with post-traumatic stress disorder and functional outcome following mild traumatic brain injury.J Clin Neurosci. 2017 Jan;35:109-116. doi: 10.1016/j.jocn.2016.09.017. Epub 2016 Oct 18. J Clin Neurosci. 2017. PMID: 27769642 Free PMC article.
-
The impact of catechol-O-methyltransferase SNPs and haplotypes on treatment response phenotypes in major depressive disorder: a case-control association study.Int Clin Psychopharmacol. 2010 Jul;25(4):218-27. doi: 10.1097/YIC.0b013e328338b884. Int Clin Psychopharmacol. 2010. PMID: 20531207
-
The Association between COMT Val158Met Polymorphism and the Post-Traumatic Stress Disorder Risk: A Meta-Analysis.Neuropsychobiology. 2022;81(2):156-170. doi: 10.1159/000514076. Epub 2021 Oct 15. Neuropsychobiology. 2022. PMID: 34657037
-
[Posttraumatic stress disorder (PTSD) as a consequence of the interaction between an individual genetic susceptibility, a traumatogenic event and a social context].Encephale. 2012 Oct;38(5):373-80. doi: 10.1016/j.encep.2011.12.003. Epub 2012 Jan 24. Encephale. 2012. PMID: 23062450 Review. French.
-
Are catechol-O-methyltransferase gene polymorphisms genetic markers for pain sensitivity after all? - A review and meta-analysis.Neurosci Biobehav Rev. 2023 May;148:105112. doi: 10.1016/j.neubiorev.2023.105112. Epub 2023 Feb 24. Neurosci Biobehav Rev. 2023. PMID: 36842714 Review.
References
-
- Amstadter AB, Nugent NR, Yang BZ, Miller A, Siburian R, Moorjani P, Haddad S, Basu A, Fagerness J, Saxe G, Smoller JW, Koenen KC, 2011. Corticotrophin-releasing hormone type 1 receptor gene (CRHR1) variants predict posttraumatic stress disorder onset and course in pediatric injury patients. Dis. Markers 30 (2–3), 89–99. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

