Bioinformatic analysis identifying FGF1 gene as a new prognostic indicator in clear cell Renal Cell Carcinoma
- PMID: 33865387
- PMCID: PMC8052755
- DOI: 10.1186/s12935-021-01917-9
Bioinformatic analysis identifying FGF1 gene as a new prognostic indicator in clear cell Renal Cell Carcinoma
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) has been the commonest renal cell carcinoma (RCC). Although the disease classification, diagnosis and targeted therapy of RCC has been increasingly evolving attributing to the rapid development of current molecular pathology, the current clinical treatment situation is still challenging considering the comprehensive and progressively developing nature of malignant cancer. The study is to identify more potential responsible genes during the development of ccRCC using bioinformatic analysis, thus aiding more precise interpretation of the disease METHODS: Firstly, different cDNA expression profiles from Gene Expression Omnibus (GEO) online database were used to screen the abnormal differently expressed genes (DEGs) between ccRCC and normal renal tissues. Then, based on the protein-protein interaction network (PPI) of all DEGs, the module analysis was performed to scale down the potential genes, and further survival analysis assisted our proceeding to the next step for selecting a credible key gene. Thirdly, immunohistochemistry (IHC) and quantitative real-time PCR (QPCR) were conducted to validate the expression change of the key gene in ccRCC comparing to normal tissues, meanwhile the prognostic value was verified using TCGA clinical data. Lastly, the potential biological function of the gene and signaling mechanism of gene regulating ccRCC development was preliminary explored.
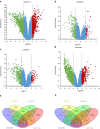
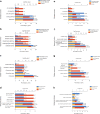
Results: Four cDNA expression profiles were picked from GEO database based on the number of containing sample cases, and a total of 192 DEGs, including 39 up-regulated and 153 down-regulated genes were shared in four profiles. Based on the DEGs PPI network, four function modules were identified highlighting a FGF1 gene involving PI3K-AKT signaling pathway which was shared in 3/4 modules. Further, both the IHC performed with ccRCC tissue microarray which contained 104 local samples and QPCR conducted using 30 different samples confirmed that FGF1 was aberrant lost in ccRCC. And Kaplan-Meier overall survival analysis revealed that FGF1 gene loss was related to worse ccRCC patients survival. Lastly, the pathological clinical features of FGF1 gene and the probable biological functions and signaling pathways it involved were analyzed using TCGA clinical data.
Conclusions: Using bioinformatic analysis, we revealed that FGF1 expression was aberrant lost in ccRCC which statistical significantly correlated with patients overall survival, and the gene's clinical features and potential biological functions were also explored. However, more detailed experiments and clinical trials are needed to support its potential drug-target role in clinical medical use.
Keywords: Clear cell renal cell carcinoma (ccRCC); FGF1 gene; GEO database; Molecular pathology; PI3K-AKT signaling pathway; Protein–protein interaction network (PPI).
Conflict of interest statement
All of the authors agreed the publication of the paper and declare no conflicts of interests.
Figures




References
-
- Williamson SR, Gill AJ, Argani P, Chen YB, Egevad L, Kristiansen G, Grignon DJ, Hes O. Report from the International Society of Urological Pathology (ISUP) Consultation Conference on Molecular Pathology of Urogenital Cancers: III: Molecular Pathology of Kidney Cancer. Am J Surg Pathol. 2020;44(7):e47–e65. doi: 10.1097/PAS.0000000000001476. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

