A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells
- PMID: 33865984
- PMCID: PMC8343164
- DOI: 10.1016/j.jtos.2021.03.010
A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells
Abstract
Purpose: Single cell (sc) analyses of key embryonic, fetal and adult stages were performed to generate a comprehensive single cell atlas of all the corneal and adjacent conjunctival cell types from development to adulthood.
Methods: Four human adult and seventeen embryonic and fetal corneas from 10 to 21 post conception week (PCW) specimens were dissociated to single cells and subjected to scRNA- and/or ATAC-Seq using the 10x Genomics platform. These were embedded using Uniform Manifold Approximation and Projection (UMAP) and clustered using Seurat graph-based clustering. Cluster identification was performed based on marker gene expression, bioinformatic data mining and immunofluorescence (IF) analysis. RNA interference, IF, colony forming efficiency and clonal assays were performed on cultured limbal epithelial cells (LECs).
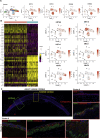
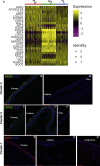
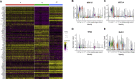
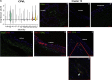
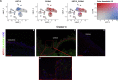
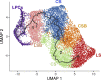
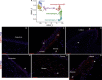
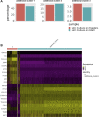
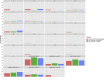
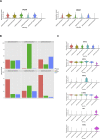
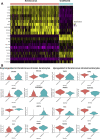
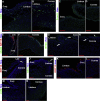
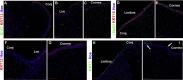
Results: scRNA-Seq analysis of 21,343 cells from four adult human corneas and adjacent conjunctivas revealed the presence of 21 cell clusters, representing the progenitor and differentiated cells in all layers of cornea and conjunctiva as well as immune cells, melanocytes, fibroblasts, and blood/lymphatic vessels. A small cell cluster with high expression of limbal progenitor cell (LPC) markers was identified and shown via pseudotime analysis to give rise to five other cell types representing all the subtypes of differentiated limbal and corneal epithelial cells. A novel putative LPCs surface marker, GPHA2, expressed on the surface of 0.41% ± 0.21 of the cultured LECs, was identified, based on predominant expression in the limbal crypts of adult and developing cornea and RNAi validation in cultured LECs. Combining scRNA- and ATAC-Seq analyses, we identified multiple upstream regulators for LPCs and demonstrated a close interaction between the immune cells and limbal progenitor cells. RNA-Seq analysis indicated the loss of GPHA2 expression and acquisition of proliferative limbal basal epithelial cell markers during ex vivo LEC expansion, independently of the culture method used. Extending the single cell analyses to keratoconus, we were able to reveal activation of collagenase in the corneal stroma and a reduced pool of limbal suprabasal cells as two key changes underlying the disease phenotype. Single cell RNA-Seq of 89,897 cells obtained from embryonic and fetal cornea indicated that during development, the conjunctival epithelium is the first to be specified from the ocular surface epithelium, followed by the corneal epithelium and the establishment of LPCs, which predate the formation of limbal niche by a few weeks.
Conclusions: Our scRNA-and ATAC-Seq data of developing and adult cornea in steady state and disease conditions provide a unique resource for defining genes/pathways that can lead to improvement in ex vivo LPCs expansion, stem cell differentiation methods and better understanding and treatment of ocular surface disorders.
Keywords: Conjunctiva; Cornea; Embryonic and fetal eye; Keratoconus; LSCs dysplasia; Limbal epithelial cells (LECs); Limbal epithelial expansion; Limbal progenitor cells (LPCs); Limbal stem cells (LSCs); Ocular surface; Single cell ATAC-Seq; Single cell RNA-Seq.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
None.
Figures













References
-
- Biomaterials and Regenerative Medicine in Ophthalmology - 1st Edition n.d. https://www.elsevier.com/books/biomaterials-and-regenerative-medicine-in... (accessed June 29, 2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

