Aminoglycosides as potential inhibitors of SARS-CoV-2 main protease: an in silico drug repurposing study on FDA-approved antiviral and anti-infection agents
- PMID: 33866129
- PMCID: PMC7871101
- DOI: 10.1016/j.jiph.2021.01.016
Aminoglycosides as potential inhibitors of SARS-CoV-2 main protease: an in silico drug repurposing study on FDA-approved antiviral and anti-infection agents
Abstract
Background: The emergence and spread of SARS-CoV-2 throughout the world has created an enormous socioeconomic impact. Although there are several promising drug candidates in clinical trials, none is available clinically. Thus, the drug repurposing approach may help to overcome the current pandemic.
Methods: The main protease (Mpro) of SARS-CoV-2 is crucial for cleaving nascent polypeptide chains. Here, FDA-approved antiviral and anti-infection drugs were screened by high-throughput virtual screening (HTVS) followed by re-docking with standard-precision (SP) and extra-precision (XP) molecular docking. The most potent drug's binding was further validated by free energy calculations (Prime/MM-GBSA) and molecular dynamics (MD) simulation.
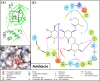
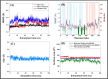
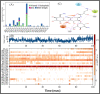
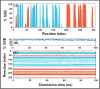
Results: Out of 1397 potential drugs, 157 showed considerable affinity toward Mpro. After HTVS, SP, and XP molecular docking, four high-affinity lead drugs (Iodixanol, Amikacin, Troxerutin, and Rutin) with docking energies -10.629 to -11.776kcal/mol range were identified. Among them, Amikacin exhibited the lowest Prime/MM-GBSA energy (-73.800kcal/mol). It led us to evaluate other aminoglycosides (Neomycin, Paramomycin, Gentamycin, Streptomycin, and Tobramycin) against Mpro. All aminoglycosides were bound to the substrate-binding site of Mpro and interacted with crucial residues. Altogether, Amikacin was found to be the most potent inhibitor of Mpro. MD simulations of the Amikacin-Mpro complex suggested the formation of a complex stabilized by hydrogen bonds, salt bridges, and van der Waals interactions.
Conclusion: Aminoglycosides may serve as a scaffold to design potent drug molecules against COVID-19. However, further validation by in vitro and in vivo studies is required before using aminoglycosides as an anti-COVID-19 agent.
Keywords: Aminoglycosides; Antibiotics; COVID-19; Docking and simulation; SARS-CoV-2.
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors report no declarations of interest.
Figures
Similar articles
-
Repurposing FDA-approved drugs for COVID-19: targeting the main protease through multi-phase in silico approach.Antivir Ther. 2024 Dec;29(6):13596535241305536. doi: 10.1177/13596535241305536. Antivir Ther. 2024. PMID: 39639531
-
Molecular insights to the binding interactions of APNS containing HIV-protease inhibitors against SARS-CoV-2 Mpro: an in silico approach towards drug repurposing.J Biomol Struct Dyn. 2023 Jun;41(9):3900-3913. doi: 10.1080/07391102.2022.2059008. Epub 2022 Apr 7. J Biomol Struct Dyn. 2023. PMID: 35388744
-
Possibility of HIV-1 protease inhibitors-clinical trial drugs as repurposed drugs for SARS-CoV-2 main protease: a molecular docking, molecular dynamics and binding free energy simulation study.J Biomol Struct Dyn. 2021 Sep;39(15):5368-5375. doi: 10.1080/07391102.2020.1786459. Epub 2020 Jul 6. J Biomol Struct Dyn. 2021. PMID: 32627689 Free PMC article.
-
In silico molecular docking, dynamics simulation and repurposing of some VEGFR-2 inhibitors based on the SARS-CoV-2-main-protease inhibitor N3.J Biomol Struct Dyn. 2023 Nov;41(19):9267-9281. doi: 10.1080/07391102.2022.2148000. Epub 2022 Nov 18. J Biomol Struct Dyn. 2023. PMID: 36399002 Review.
-
Haste makes waste: A critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition.Med Res Rev. 2022 Mar;42(2):744-769. doi: 10.1002/med.21862. Epub 2021 Oct 26. Med Res Rev. 2022. PMID: 34697818 Free PMC article. Review.
Cited by
-
Novel Drug Design for Treatment of COVID-19: A Systematic Review of Preclinical Studies.Can J Infect Dis Med Microbiol. 2022 Sep 25;2022:2044282. doi: 10.1155/2022/2044282. eCollection 2022. Can J Infect Dis Med Microbiol. 2022. PMID: 36199815 Free PMC article. Review.
-
Identification of novel transmembrane Protease Serine Type 2 drug candidates for COVID-19 using computational studies.Inform Med Unlocked. 2021;26:100725. doi: 10.1016/j.imu.2021.100725. Epub 2021 Sep 7. Inform Med Unlocked. 2021. PMID: 34514079 Free PMC article.
-
An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2.Comput Struct Biotechnol J. 2021;19:4868-4883. doi: 10.1016/j.csbj.2021.08.036. Epub 2021 Aug 24. Comput Struct Biotechnol J. 2021. PMID: 34457214 Free PMC article. Review.
-
Targeting SARS-CoV-2 main protease by teicoplanin: A mechanistic insight by docking, MM/GBSA and molecular dynamics simulation.J Mol Struct. 2021 Dec 15;1246:131124. doi: 10.1016/j.molstruc.2021.131124. Epub 2021 Jul 18. J Mol Struct. 2021. PMID: 34305175 Free PMC article.
-
COVID-19-specific transcriptomic signature detectable in blood across multiple cohorts.Front Genet. 2022 Aug 5;13:929887. doi: 10.3389/fgene.2022.929887. eCollection 2022. Front Genet. 2022. PMID: 35991542 Free PMC article.
References
-
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

