Deep Learning-Based Classification of GAD67-Positive Neurons Without the Immunosignal
- PMID: 33867947
- PMCID: PMC8044854
- DOI: 10.3389/fnana.2021.643067
Deep Learning-Based Classification of GAD67-Positive Neurons Without the Immunosignal
Abstract
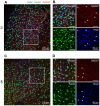
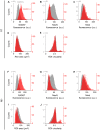
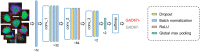
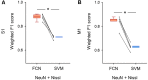
Excitatory neurons and GABAergic interneurons constitute neural circuits and play important roles in information processing. In certain brain regions, such as the neocortex and the hippocampus, there are fewer interneurons than excitatory neurons. Interneurons have been quantified via immunohistochemistry, for example, for GAD67, an isoform of glutamic acid decarboxylase. Additionally, the expression level of other proteins varies among cell types. For example, NeuN, a commonly used marker protein for postmitotic neurons, is expressed differently across brain regions and cell classes. Thus, we asked whether GAD67-immunopositive neurons can be detected using the immunofluorescence signals of NeuN and the fluorescence signals of Nissl substances. To address this question, we stained neurons in layers 2/3 of the primary somatosensory cortex (S1) and the primary motor cortex (M1) of mice and manually labeled the neurons as either cell type using GAD67 immunosignals. We then sought to detect GAD67-positive neurons without GAD67 immunosignals using a custom-made deep learning-based algorithm. Using this deep learning-based model, we succeeded in the binary classification of the neurons using Nissl and NeuN signals without referring to the GAD67 signals. Furthermore, we confirmed that our deep learning-based method surpassed classic machine-learning methods in terms of binary classification performance. Combined with the visualization of the hidden layer of our deep learning algorithm, our model provides a new platform for identifying unbiased criteria for cell-type classification.
Keywords: GAD67; NeuN; deep learning; fully convolutional network; interneuron; motor cortex; mouse; somatosensory cortex.
Copyright © 2021 Yamashiro, Liu, Matsumoto and Ikegaya.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

