Holstein and Jersey Steers Differ in Rumen Microbiota and Enteric Methane Emissions Even Fed the Same Total Mixed Ration
- PMID: 33868186
- PMCID: PMC8044996
- DOI: 10.3389/fmicb.2021.601061
Holstein and Jersey Steers Differ in Rumen Microbiota and Enteric Methane Emissions Even Fed the Same Total Mixed Ration
Abstract
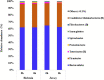
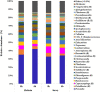
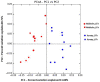
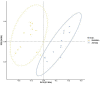
Previous studies have focused on the rumen microbiome and enteric methane (CH4) emissions in dairy cows, yet little is known about steers, especially steers of dairy breeds. In the present study, we comparatively examined the rumen microbiota, fermentation characteristics, and CH4 emissions from six non-cannulated Holstein (710.33 ± 43.02 kg) and six Jersey (559.67 ± 32.72 kg) steers. The steers were fed the same total mixed ration (TMR) for 30 days. After 25 days of adaptation to the diet, CH4 emissions were measured using GreenFeed for three consecutive days, and rumen fluid samples were collected on last day using stomach tubing before feeding (0 h) and 6 h after feeding. CH4 production (g/d/animal), CH4 yield (g/kg DMI), and CH4 intensity (g/kg BW0.75) were higher in the Jersey steers than in the Holstein steers. The lowest pH value was recorded at 6 h after feeding. The Jersey steers had lower rumen pH and a higher concentration of ammonia-nitrogen (NH3-N). The Jersey steers had a numerically higher molar proportion of acetate than the Holstein steers, but the opposite was true for that of propionate. Metataxonomic analysis of the rumen microbiota showed that the two breeds had similar species richness, Shannon, and inverse Simpson diversity indexes. Principal coordinates analysis showed that the overall rumen microbiota was different between the two breeds. Both breeds were dominated by Prevotella ruminicola, and its highest relative abundance was observed 6 h after feeding. The genera Ethanoligenens, Succinivibrio, and the species Ethanoligenens harbinense, Succinivibrio dextrinosolvens, Prevotella micans, Prevotella copri, Prevotella oris, Prevotella baroniae, and Treponema succinifaciens were more abundant in Holstein steers while the genera Capnocytophaga, Lachnoclostridium, Barnesiella, Oscillibacter, Galbibacter, and the species Capnocytophaga cynodegmi, Galbibacter mesophilus, Barnesiella intestinihominis, Prevotella shahii, and Oscillibacter ruminantium in the Jersey steers. The Jersey steers were dominated by Methanobrevibacter millerae while the Holstein steers by Methanobrevibacter olleyae. The overall results suggest that sampling hour has little influence on the rumen microbiota; however, breeds of steers can affect the assemblage of the rumen microbiota and different mitigation strategies may be needed to effectively manipulate the rumen microbiota and mitigate enteric CH4 emissions from these steers.
Keywords: Holstein steer; Jersey steer; enteric methane emissions; rumen fermentation; rumen microbiota.
Copyright © 2021 Islam, Kim, Ramos, Mamuad, Son, Yu, Lee, Cho and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Aoac. (2005). Official methods of analysis of the Association of Official Analytical Chemists. Gaithersburg: AOAC International.
-
- Bainbridge M. L., Cersosimo L. M., Wright A. D. G., Kraft J. (2016). Rumen bacterial communities shift across a lactation in holstein, jersey and holstein × jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 92 1–14. 10.1093/femsec/fiw059 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

