Recurrent Potential G-Quadruplex Sequences in Archaeal Genomes
- PMID: 33868206
- PMCID: PMC8044849
- DOI: 10.3389/fmicb.2021.647851
Recurrent Potential G-Quadruplex Sequences in Archaeal Genomes
Abstract
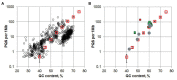
Evolutionary conservation or over-representation of the potential G-quadruplex sequences (PQS) in genomes are usually considered as a sign of the functional relevance of these sequences. However, uneven base distribution (GC-content) along the genome may along the genome may result in seeming abundance of PQSs over average in the genome. Apart from this, a number of other conserved functional signals that are encoded in the GC-rich genomic regions may inadvertently result in emergence of G-quadruplex compatible sequences. Here, we analyze the genomes of archaea focusing our search to repetitive PQS (rPQS) motifs within each organism. The probability of occurrence of several identical PQSs within a relatively short archaeal genome is low and, thus, the structure and genomic location of such rPQSs may become a direct indication of their functionality. We have found that the majority of the genomes of Methanomicrobiaceae family of archaea contained multiple copies of the interspersed highly similar PQSs. Short oligonucleotides corresponding to the rPQS formed the G-quadruplex (G4) structure in presence of potassium ions as demonstrated by circular dichroism (CD) and enzymatic probing. However, further analysis of the genomic context for the rPQS revealed a 10-12 nt cytosine-rich track adjacent to 3'-end of each rPQS. Synthetic DNA fragments that included the C-rich track tended to fold into alternative structures such as hairpin structure and antiparallel triplex that were in equilibrium with G4 structure depending on the presence of potassium ions in solution. Structural properties of the found repetitive sequences, their location in the genomes of archaea, and possible functions are discussed.
Keywords: DNA; G-quadruplex; Methanomicrobiaceae; archaea; circular dichroism; nuclease probing.
Copyright © 2021 Chashchina, Shchyolkina, Kolosov, Beniaminov and Kaluzhny.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Case studies on potential G-quadruplex-forming sequences from the bacterial orders Deinococcales and Thermales derived from a survey of published genomes.Sci Rep. 2018 Oct 24;8(1):15679. doi: 10.1038/s41598-018-33944-4. Sci Rep. 2018. PMID: 30356061 Free PMC article.
-
Presence, Location and Conservation of Putative G-Quadruplex Forming Sequences in Arboviruses Infecting Humans.Int J Mol Sci. 2023 May 30;24(11):9523. doi: 10.3390/ijms24119523. Int J Mol Sci. 2023. PMID: 37298474 Free PMC article.
-
Human DNA Repair Genes Possess Potential G-Quadruplex Sequences in Their Promoters and 5'-Untranslated Regions.Biochemistry. 2018 Feb 13;57(6):991-1002. doi: 10.1021/acs.biochem.7b01172. Epub 2018 Jan 24. Biochemistry. 2018. PMID: 29320161 Free PMC article.
-
Dual role of G-quadruplex in translocation renal cell carcinoma: Exploring plausible Cancer therapeutic innovation.Biochim Biophys Acta Gen Subj. 2020 Dec;1864(12):129719. doi: 10.1016/j.bbagen.2020.129719. Epub 2020 Aug 31. Biochim Biophys Acta Gen Subj. 2020. PMID: 32882363 Review.
-
Colocalization of m6A and G-Quadruplex-Forming Sequences in Viral RNA (HIV, Zika, Hepatitis B, and SV40) Suggests Topological Control of Adenosine N 6-Methylation.ACS Cent Sci. 2019 Feb 27;5(2):218-228. doi: 10.1021/acscentsci.8b00963. Epub 2019 Feb 4. ACS Cent Sci. 2019. PMID: 30834310 Free PMC article. Review.
Cited by
-
DNA Methylation: Genomewide Distribution, Regulatory Mechanism and Therapy Target.Acta Naturae. 2022 Oct-Dec;14(4):4-19. doi: 10.32607/actanaturae.11822. Acta Naturae. 2022. PMID: 36694897 Free PMC article.
-
G-quadruplex propensity in H. neanderthalensis, H. sapiens and Denisovans mitochondrial genomes.NAR Genom Bioinform. 2024 May 30;6(2):lqae060. doi: 10.1093/nargab/lqae060. eCollection 2024 Jun. NAR Genom Bioinform. 2024. PMID: 38817800 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

