Antiviral Activity of Interferon Alpha-Inducible Protein 27 Against Hepatitis B Virus Gene Expression and Replication
- PMID: 33868214
- PMCID: PMC8044325
- DOI: 10.3389/fmicb.2021.656353
Antiviral Activity of Interferon Alpha-Inducible Protein 27 Against Hepatitis B Virus Gene Expression and Replication
Abstract
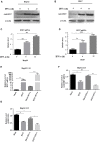
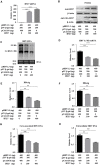
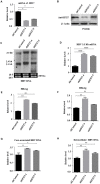
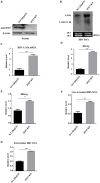
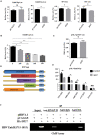
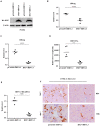
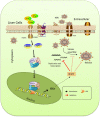
Despite the availability of effective vaccines, hepatitis B virus (HBV) is still a major health issue, and approximately 350 million people have been chronically infected with HBV throughout the world. Interferons (IFNs) are the key molecules in the innate immune response that restrict several kinds of viral infections via the induction of hundreds of IFN-stimulated genes (ISGs). The objective of this study was to confirm if interferon alpha-inducible protein 27 (IFI27) as an ISG could inhibit HBV gene expression and DNA replication both in cell culture and in a mouse model. In human hepatoma cells, IFI27 was highly induced by the stimulation of IFN-alpha (IFN-α), and it potentiated the anti-HBV activity. The overexpression of IFI27 inhibited, while its silencing enhanced the HBV replication in HepG2 cell. However, the knocking out of IFI27 in HepG2 cells robustly increases the formation of viral DNA, RNA, and proteins. Detailed mechanistic analysis of the HBV genome showed that a sequence [nucleotide (nt) 1715-1815] of the EnhII/Cp promoter was solely responsible for viral inhibition. Similarly, the hydrodynamic injection of IFI27 expression constructs along with the HBV genome into mice resulted in a significant reduction in viral gene expression and DNA replication. In summary, our studies suggested that IFI27 contributed a vital role in HBV gene expression and replication and IFI27 may be a potential antiviral agent for the treatment of HBV.
Keywords: EnhII/Cp; IFI27; ISGs; antiviral activity; hepatitis B virus.
Copyright © 2021 Ullah, Sajid, Yan, Feng, He, Shereen, Li, Xu, Hao, Guo, Chen, Zhou and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Interferon and interferon-stimulated genes in HBV treatment.Front Immunol. 2022 Dec 1;13:1034968. doi: 10.3389/fimmu.2022.1034968. eCollection 2022. Front Immunol. 2022. PMID: 36531993 Free PMC article. Review.
-
The Functional and Antiviral Activity of Interferon Alpha-Inducible IFI6 Against Hepatitis B Virus Replication and Gene Expression.Front Immunol. 2021 Apr 1;12:634937. doi: 10.3389/fimmu.2021.634937. eCollection 2021. Front Immunol. 2021. PMID: 33868257 Free PMC article.
-
SOX9 represses hepatitis B virus replication through binding to HBV EnhII/Cp and inhibiting the promoter activity.Antiviral Res. 2020 May;177:104761. doi: 10.1016/j.antiviral.2020.104761. Epub 2020 Mar 5. Antiviral Res. 2020. PMID: 32147495
-
Interferon-inducible MX2 is a host restriction factor of hepatitis B virus replication.J Hepatol. 2020 May;72(5):865-876. doi: 10.1016/j.jhep.2019.12.009. Epub 2019 Dec 18. J Hepatol. 2020. PMID: 31863794
-
Intracellular interferon signalling pathways as potential regulators of covalently closed circular DNA in the treatment of chronic hepatitis B.World J Gastroenterol. 2021 Apr 14;27(14):1369-1391. doi: 10.3748/wjg.v27.i14.1369. World J Gastroenterol. 2021. PMID: 33911462 Free PMC article. Review.
Cited by
-
Novel Biomarkers of Hepatitis B Virus and Their Use in Chronic Hepatitis B Patient Management.Viruses. 2021 May 21;13(6):951. doi: 10.3390/v13060951. Viruses. 2021. PMID: 34064049 Free PMC article. Review.
-
Whole blood transcriptome analysis in dairy calves experimentally challenged with bovine herpesvirus 1 (BoHV-1) and comparison to a bovine respiratory syncytial virus (BRSV) challenge.Front Genet. 2023 Feb 17;14:1092877. doi: 10.3389/fgene.2023.1092877. eCollection 2023. Front Genet. 2023. PMID: 36873940 Free PMC article.
-
Insight Into the Long Noncoding RNA and mRNA Coexpression Profile in the Human Blood Transcriptome Upon Leishmania infantum Infection.Front Immunol. 2022 Mar 15;13:784463. doi: 10.3389/fimmu.2022.784463. eCollection 2022. Front Immunol. 2022. PMID: 35370994 Free PMC article.
-
Interferon and interferon-stimulated genes in HBV treatment.Front Immunol. 2022 Dec 1;13:1034968. doi: 10.3389/fimmu.2022.1034968. eCollection 2022. Front Immunol. 2022. PMID: 36531993 Free PMC article. Review.
-
Identification of dynamic gene expression profiles during sequential vaccination with ChAdOx1/BNT162b2 using machine learning methods.Front Microbiol. 2023 Mar 17;14:1138674. doi: 10.3389/fmicb.2023.1138674. eCollection 2023. Front Microbiol. 2023. PMID: 37007526 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

