Oncogenic mechanism-based pharmaceutical validation of therapeutics targeting MET receptor tyrosine kinase
- PMID: 33868463
- PMCID: PMC8020248
- DOI: 10.1177/17588359211006957
Oncogenic mechanism-based pharmaceutical validation of therapeutics targeting MET receptor tyrosine kinase
Abstract
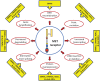
Aberrant expression and/or activation of the MET receptor tyrosine kinase is characterized by genomic recombination, gene amplification, activating mutation, alternative exon-splicing, increased transcription, and their different combinations. These dysregulations serve as oncogenic determinants contributing to cancerous initiation, progression, malignancy, and stemness. Moreover, integration of the MET pathway into the cellular signaling network as an addiction mechanism for survival has made this receptor an attractive pharmaceutical target for oncological intervention. For the last 20 years, MET-targeting small-molecule kinase inhibitors (SMKIs), conventional therapeutic monoclonal antibodies (TMABs), and antibody-based biotherapeutics such as bispecific antibodies, antibody-drug conjugates (ADC), and dual-targeting ADCs have been under intensive investigation. Outcomes from preclinical studies and clinical trials are mixed with certain successes but also various setbacks. Due to the complex nature of MET dysregulation with multiple facets and underlying mechanisms, mechanism-based validation of MET-targeting therapeutics is crucial for the selection and validation of lead candidates for clinical trials. In this review, we discuss the importance of various types of mechanism-based pharmaceutical models in evaluation of different types of MET-targeting therapeutics. The advantages and disadvantages of these mechanism-based strategies for SMKIs, conventional TMABs, and antibody-based biotherapeutics are analyzed. The demand for establishing new strategies suitable for validating novel biotherapeutics is also discussed. The information summarized should provide a pharmaceutical guideline for selection and validation of MET-targeting therapeutics for clinical application in the future.
Keywords: MET receptor tyrosine kinase; antibody–drug conjugates; bispecific antibody; dual-targeting ADC; pharmaceutical validation; small-molecule kinase inhibitor; therapeutic monoclonal antibody; tumorigenic mechanism.
© The Author(s), 2021.
Conflict of interest statement
Conflict of interest statement: The authors declare that there is no conflict of interest.
Figures



References
-
- Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984; 311: 29–33. - PubMed
-
- Malik R, Mambetsariev I, Fricke J, et al. MET receptor in oncology: from biomarker to therapeutic target. Adv Cancer Res 2020; 147: 259–301. - PubMed
-
- Onzetto C, Bardelli A, Zhen Z, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 1994; 77: 261–271. - PubMed
-
- Peschard P, Fournier TM, Lamorte L, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell 2001; 8: 995–1004. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

