Ancestral sequence reconstruction - An underused approach to understand the evolution of gene function in plants?
- PMID: 33868595
- PMCID: PMC8039532
- DOI: 10.1016/j.csbj.2021.03.008
Ancestral sequence reconstruction - An underused approach to understand the evolution of gene function in plants?
Abstract
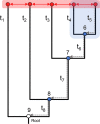
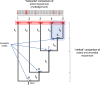
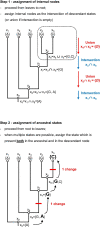
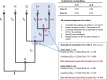
Whilst substantial research effort has been placed on understanding the interactions of plant proteins with their molecular partners, relatively few studies in plants - by contrast to work in other organisms - address how these interactions evolve. It is thought that ancestral proteins were more promiscuous than modern proteins and that specificity often evolved following gene duplication and subsequent functional refining. However, ancestral protein resurrection studies have found that some modern proteins have evolved de novo from ancestors lacking those functions. Intriguingly, the new interactions evolved as a consequence of just a few mutations and, as such, acquisition of new functions appears to be neither difficult nor rare, however, only a few of them are incorporated into biological processes before they are lost to subsequent mutations. Here, we detail the approach of ancestral sequence reconstruction (ASR), providing a primer to reconstruct the sequence of an ancestral gene. We will present case studies from a range of different eukaryotes before discussing the few instances where ancestral reconstructions have been used in plants. As ASR is used to dig into the remote evolutionary past, we will also present some alternative genetic approaches to investigate molecular evolution on shorter timescales. We argue that the study of plant secondary metabolism is particularly well suited for ancestral reconstruction studies. Indeed, its ancient evolutionary roots and highly diverse landscape provide an ideal context in which to address the focal issue around the emergence of evolutionary novelties and how this affects the chemical diversification of plant metabolism.
Keywords: APR, ancestral protein resurrection; ASR, ancestral sequence reconstruction; Ancestral sequence reconstruction; CDS, coding sequence; Evolution; GR, glucocorticoid receptor; GWAS, genome wide association study; Genomics; InDel, insertion/deletion; MCMC, Markov Chain Monte Carlo; ML, maximum likelihood; MP, maximum parsimony; MR, mineralcorticoid receptor; MSA, multiple sequence alignment; Metabolism; NJ, neighbor-joining; Phylogenetics; Plants; SFS, site frequency spectrum.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition.Elife. 2022 Nov 15;11:e81050. doi: 10.7554/eLife.81050. Elife. 2022. PMID: 36377784 Free PMC article.
-
Applications of ancestral sequence reconstruction for understanding the evolution of plant specialized metabolism.Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230348. doi: 10.1098/rstb.2023.0348. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343033 Free PMC article. Review.
-
Evolution of protein specificity: insights from ancestral protein reconstruction.Curr Opin Struct Biol. 2017 Dec;47:113-122. doi: 10.1016/j.sbi.2017.07.003. Epub 2017 Aug 23. Curr Opin Struct Biol. 2017. PMID: 28841430 Free PMC article. Review.
-
Contingency and chance erase necessity in the experimental evolution of ancestral proteins.Elife. 2021 Jun 1;10:e67336. doi: 10.7554/eLife.67336. Elife. 2021. PMID: 34061027 Free PMC article.
-
Using the Evolutionary History of Proteins to Engineer Insertion-Deletion Mutants from Robust, Ancestral Templates Using Graphical Representation of Ancestral Sequence Predictions (GRASP).Methods Mol Biol. 2022;2397:85-110. doi: 10.1007/978-1-0716-1826-4_6. Methods Mol Biol. 2022. PMID: 34813061
Cited by
-
Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition.Elife. 2022 Nov 15;11:e81050. doi: 10.7554/eLife.81050. Elife. 2022. PMID: 36377784 Free PMC article.
-
Natural variation of respiration-related traits in plants.Plant Physiol. 2023 Apr 3;191(4):2120-2132. doi: 10.1093/plphys/kiac593. Plant Physiol. 2023. PMID: 36546766 Free PMC article. Review.
-
Ancestral Sequence Reconstruction for Designing Biocatalysts and Investigating their Functional Mechanisms.JACS Au. 2024 Oct 25;4(12):4571-4591. doi: 10.1021/jacsau.4c00653. eCollection 2024 Dec 23. JACS Au. 2024. PMID: 39735918 Free PMC article. Review.
-
Zebrafish: unraveling genetic complexity through duplicated genes.Dev Genes Evol. 2024 Dec;234(2):99-116. doi: 10.1007/s00427-024-00720-6. Epub 2024 Jul 30. Dev Genes Evol. 2024. PMID: 39079985 Free PMC article. Review.
-
Applications of ancestral sequence reconstruction for understanding the evolution of plant specialized metabolism.Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230348. doi: 10.1098/rstb.2023.0348. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343033 Free PMC article. Review.
References
-
- Tautz D., Ellegren H., Weigel D. Next generation molecular ecology. Mol Ecol. 2010;19(Suppl 1):1–3. - PubMed
-
- Exposito-Alonso M., Drost H.G., Burbano H.A., Weigel D. The Earth BioGenome project: opportunities and challenges for plant genomics and conservation. Plant J. 2020;102:222–229. - PubMed
-
- Thornton J.W. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–375. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
