Human ACE2-Functionalized Gold "Virus-Trap" Nanostructures for Accurate Capture of SARS-CoV-2 and Single-Virus SERS Detection
- PMID: 33868761
- PMCID: PMC8042470
- DOI: 10.1007/s40820-021-00620-8
Human ACE2-Functionalized Gold "Virus-Trap" Nanostructures for Accurate Capture of SARS-CoV-2 and Single-Virus SERS Detection
Abstract
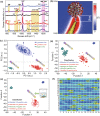
The current COVID-19 pandemic urges the extremely sensitive and prompt detection of SARS-CoV-2 virus. Here, we present a Human Angiotensin-converting-enzyme 2 (ACE2)-functionalized gold "virus traps" nanostructure as an extremely sensitive SERS biosensor, to selectively capture and rapidly detect S-protein expressed coronavirus, such as the current SARS-CoV-2 in the contaminated water, down to the single-virus level. Such a SERS sensor features extraordinary 106-fold virus enrichment originating from high-affinity of ACE2 with S protein as well as "virus-traps" composed of oblique gold nanoneedles, and 109-fold enhancement of Raman signals originating from multi-component SERS effects. Furthermore, the identification standard of virus signals is established by machine-learning and identification techniques, resulting in an especially low detection limit of 80 copies mL-1 for the simulated contaminated water by SARS-CoV-2 virus with complex circumstance as short as 5 min, which is of great significance for achieving real-time monitoring and early warning of coronavirus. Moreover, here-developed method can be used to establish the identification standard for future unknown coronavirus, and immediately enable extremely sensitive and rapid detection of novel virus.
Supplementary information: The online version contains supplementary material available at 10.1007/s40820-021-00620-8.
Keywords: Human ACE2; SARS-CoV-2; SERS; Single-virus detection; “Virus-trap” nanostructure.
© The Author(s) 2021.
Figures


References
-
- WHO Coronavirus Disease (COVID-19) Dashboard, data last updated: 2020/12/1, 7:08pm CEST. https://covid19.who.int/ (2020)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
