Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture
- PMID: 33868800
- PMCID: PMC8035907
- DOI: 10.7717/peerj.10941
Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture
Abstract
Background: The chicken is the most abundant food animal in the world. However, despite its importance, the chicken gut microbiome remains largely undefined. Here, we exploit culture-independent and culture-dependent approaches to reveal extensive taxonomic diversity within this complex microbial community.
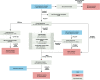
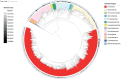
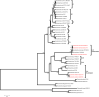
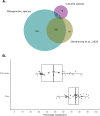
Results: We performed metagenomic sequencing of fifty chicken faecal samples from two breeds and analysed these, alongside all (n = 582) relevant publicly available chicken metagenomes, to cluster over 20 million non-redundant genes and to construct over 5,500 metagenome-assembled bacterial genomes. In addition, we recovered nearly 600 bacteriophage genomes. This represents the most comprehensive view of taxonomic diversity within the chicken gut microbiome to date, encompassing hundreds of novel candidate bacterial genera and species. To provide a stable, clear and memorable nomenclature for novel species, we devised a scalable combinatorial system for the creation of hundreds of well-formed Latin binomials. We cultured and genome-sequenced bacterial isolates from chicken faeces, documenting over forty novel species, together with three species from the genus Escherichia, including the newly named species Escherichia whittamii.
Conclusions: Our metagenomic and culture-based analyses provide new insights into the bacterial, archaeal and bacteriophage components of the chicken gut microbiome. The resulting datasets expand the known diversity of the chicken gut microbiome and provide a key resource for future high-resolution taxonomic and functional studies on the chicken gut microbiome.
Keywords: Bacterial nomenclature; Biodiversity; Candidatus; Chickens; Gut microbiome; Metagenome-assembled genome; Metagenomics; Uncultured bacteria.
© 2021 Gilroy et al.
Conflict of interest statement
Arss Secka is employed by the West Africa Livestock Innovation Centre.
Figures


References
-
- Amoozegar MA, Bagheri M, Makhdoumi-Kakhki A, Didari M, Schumann P, Nikou MM, Sánchez-Porro C, Ventosa A. Aliicoccus persicus gen. nov., sp. nov., a halophilic member of the Firmicutes isolated from a hypersaline lake. International Journal of Systematic and Evolutionary Microbiology. 2014;64(Pt_6):1964–1969. doi: 10.1099/ijs.0.058545-0. - DOI - PubMed
Associated data
Grants and funding
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10355/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10356/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L015080/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

