A Nomogram Combining a Four-Gene Biomarker and Clinical Factors for Predicting Survival of Melanoma
- PMID: 33868993
- PMCID: PMC8047639
- DOI: 10.3389/fonc.2021.593587
A Nomogram Combining a Four-Gene Biomarker and Clinical Factors for Predicting Survival of Melanoma
Abstract
Background: Currently there is no effective prognostic indicator for melanoma, the deadliest skin cancer. Thus, we aimed to develop and validate a nomogram predictive model for predicting survival of melanoma.
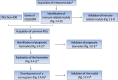
Methods: Four hundred forty-nine melanoma cases with RNA sequencing (RNA-seq) data from TCGA were randomly divided into the training set I (n = 224) and validation set I (n = 225), 210 melanoma cases with RNA-seq data from Lund cohort of Lund University (available in GSE65904) were used as an external test set. The prognostic gene biomarker was developed and validated based on the above three sets. The developed gene biomarker combined with clinical characteristics was used as variables to develop and validate a nomogram predictive model based on 379 patients with complete clinical data from TCGA (Among 470 cases, 91 cases with missing clinical data were excluded from the study), which were randomly divided into the training set II (n = 189) and validation set II (n = 190). Area under the curve (AUC), concordance index (C-index), calibration curve, and Kaplan-Meier estimate were used to assess predictive performance of the nomogram model.
Results: Four genes, i.e., CLEC7A, CLEC10A, HAPLN3, and HCP5 comprise an immune-related prognostic biomarker. The predictive performance of the biomarker was validated using tROC and log-rank test in the training set I (n = 224, 5-year AUC of 0.683), validation set I (n = 225, 5-year AUC of 0.644), and test set I (n = 210, 5-year AUC of 0.645). The biomarker was also significantly associated with improved survival in the training set (P < 0.01), validation set (P < 0.05), and test set (P < 0.001), respectively. In addition, a nomogram combing the four-gene biomarker and six clinical factors for predicting survival in melanoma was developed in the training set II (n = 189), and validated in the validation set II (n = 190), with a concordance index of 0.736 ± 0.041 and an AUC of 0.832 ± 0.071.
Conclusion: We developed and validated a nomogram predictive model combining a four-gene biomarker and six clinical factors for melanoma patients, which could facilitate risk stratification and treatment planning.
Keywords: immune genes; melanoma; microenvironment; nomogram; prognostic biomarker.
Copyright © 2021 Zhang, Dang, Wang and Cong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo (2014) 28:1005–11. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

