Deciphering Gut Microbiota Dysbiosis and Corresponding Genetic and Metabolic Dysregulation in Psoriasis Patients Using Metagenomics Sequencing
- PMID: 33869074
- PMCID: PMC8047475
- DOI: 10.3389/fcimb.2021.605825
Deciphering Gut Microbiota Dysbiosis and Corresponding Genetic and Metabolic Dysregulation in Psoriasis Patients Using Metagenomics Sequencing
Abstract
Background: Increasing evidence has shown that alterations in the intestinal microbiota play an important role in the pathogenesis of psoriasis. The existing relevant studies focus on 16S rRNA gene sequencing, but in-depth research on gene functions and comprehensive identification of microbiota is lacking.
Objectives: To comprehensively identify characteristic gut microbial compositions, genetic functions and relative metabolites of patients with psoriasis and to reveal the potential pathogenesis of psoriasis.
Methods: DNA was extracted from the faecal microbiota of 30 psoriatic patients and 15 healthy subjects, and metagenomics sequencing and bioinformatic analyses were performed. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database, cluster of orthologous groups (COG) annotations, and metabolic analyses were used to indicate relative target genes and pathways to reveal the pathogenesis of psoriasis.
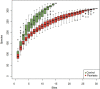
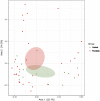
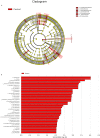
Results: Compared with healthy individuals, the gut microbiota of psoriasis patients displayed an alteration in microbial taxa distribution, but no significant difference in microbial diversity. A distinct gut microbial composition in patients with psoriasis was observed, with an increased abundance of the phyla Firmicutes, Actinobacteria and Verrucomicrobia and genera Faecalibacterium, Bacteroides, Bifidobacterium, Megamonas and Roseburia and a decreased abundance of the phyla Bacteroidetes, Euryarchaeota and Proteobacteria and genera Prevotella, Alistipes, and Eubacterium. A total of 134 COGs were predicted with functional analysis, and 15 KEGG pathways, including lipopolysaccharide (LPS) biosynthesis, WNT signaling, apoptosis, bacterial secretion system, and phosphotransferase system, were significantly enriched in psoriasis patients. Five metabolites, hydrogen sulfide (H2S), isovalerate, isobutyrate, hyaluronan and hemicellulose, were significantly dysregulated in the psoriatic cohort. The dysbiosis of gut microbiota, enriched pathways and dysregulated metabolites are relevant to immune and inflammatory response, apoptosis, the vascular endothelial growth factor (VEGF) signaling pathway, gut-brain axis and brain-skin axis that play important roles in the pathogenesis of psoriasis.
Conclusions: A clear dysbiosis was displayed in the gut microbiota profile, genetic functions and relative metabolites of psoriasis patients. This study is beneficial for further understanding the inflammatory pathogenesis of psoriasis and could be used to develop microbiome-based predictions and therapeutic approaches.
Keywords: genetic functions; gut microbiota; metabolites; metagenomics sequencing; psoriasis.
Copyright © 2021 Xiao, Zhang, Jiang, Liu, Wang, Li, Cheng, Lv, Xian, Guo and Tan.
Conflict of interest statement
MC was employed by the company Beijing QuantiHealth Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Bray J. R., Curtis J. T. (1957). An ordination of the upland forest communities of Southern Wisconsin. Eco. Monogr. 27, 325–349. 10.2307/1942268 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

