Urea as a By-Product of Ammonia Metabolism Can Be a Potential Serum Biomarker of Hepatocellular Carcinoma
- PMID: 33869206
- PMCID: PMC8047217
- DOI: 10.3389/fcell.2021.650748
Urea as a By-Product of Ammonia Metabolism Can Be a Potential Serum Biomarker of Hepatocellular Carcinoma
Abstract
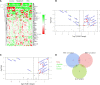
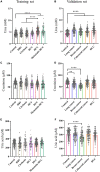
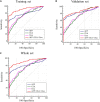
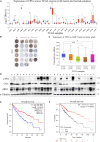
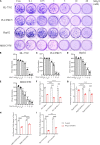
Hepatocellular carcinoma (HCC) is highly malignant; nearly half of the new cases and deaths are in China. The poor prognosis of HCC is mainly due to late diagnosis; many new biomarkers have been developed for HCC diagnosis. However, few markers are quickly translated into clinical practice; early and differential diagnosis of HCC from cirrhosis and/or hepatitis is still a clinical challenge. Metabolomics and biochemical methods were used to reveal specific serum biomarkers of HCC. Most of the elevated metabolites in HCC and HBV patients were overlapped compared with controls. Urea was the specifically elevated serum biomarker of HCC patients. Moreover, urea combined with AFP and CEA can improve the sensitivity of HCC diagnosis. The plasma ammonia of HCC patients was significantly higher than healthy controls. Co-culture cell model revealed normal liver cells cooperated with cancer cells to metabolize ammonia into urea. The urea metabolism in cancer cells marginally depended on the expression of CPS1. However, the expression of CPS1 did not change with ammonium chloride, which might regulate the urea cycle through enzyme activity. The urea cycle could detoxify high concentrations of ammonia to promote cancer cell proliferation. Therefore, urea was a by-product of ammonia metabolism and could be a potential serum biomarker for HCC. The combined application of metabolomics and biochemical methods can discover new biomarkers for the early diagnosis of HCC and be quickly applied to clinical diagnosis.
Keywords: CPS1; ammonia; biomarker; hepatocellular carcinoma; metabolomics; urea.
Copyright © 2021 Bai, Wang, Dong, Li, Yu, Guo, Zhou, Li, Yan, Wang, Wang, Li and Ren.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Ahn J. C., Teng P. C., Chen P. J., Posadas E., Tseng H. R., Lu S. C., et al. (2020). Detection of circulating tumor cells and their implications as a novel biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology 73 422–436. 10.1002/hep.31165 - DOI - PMC - PubMed
-
- Braissant O. (2010). Ammonia toxicity to the brain: effects on creatine metabolism and transport and protective roles of creatine. Mol. Genet. Metab. 100(Suppl. 1) S53–S58. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

