M-Cadherin Is a PAX3 Target During Myotome Patterning
- PMID: 33869209
- PMCID: PMC8047199
- DOI: 10.3389/fcell.2021.652652
M-Cadherin Is a PAX3 Target During Myotome Patterning
Abstract
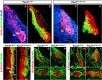
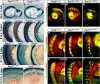
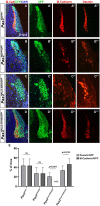
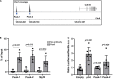
PAX3 belongs to the paired-homeobox family of transcription factors and plays a key role as an upstream regulator of muscle progenitor cells during embryonic development. Pax3-mutant embryos display impaired somite development, yet the consequences for myotome formation have not been characterized. The early myotome is formed by PAX3-expressing myogenic cells that delaminate from the dermomyotomal lips and migrate between the dermomyotome and sclerotome where they terminally differentiate. Here we show that in Pax3-mutant embryos, myotome formation is impaired, displays a defective basal lamina and the regionalization of the structural protein Desmin is lost. In addition, this phenotype is more severe in embryos combining Pax3-null and Pax3 dominant-negative alleles. We identify the adhesion molecule M-Cadherin as a PAX3 target gene, the expression of which is modulated in the myotome according to Pax3 gain- and loss-of-function alleles analyzed. Taken together, we identify M-Cadherin as a PAX3-target linked to the formation of the myotome.
Keywords: CDH15; M-Cadherin; PAX3; myogenesis; myotome.
Copyright © 2021 Esteves de Lima, Bou Akar, Mansour, Rocancourt, Buckingham and Relaix.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Auerbach R. (1954). Analysis of the developmental effects of a lethal mutation in the house mouse. J. Exp. Zool. 127 305–329. 10.1002/jez.1401270206 - DOI
-
- Bober E., Franz T., Arnold H. H., Gruss P., Tremblay P. (1994). Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Dev. Camb. Engl. 120 603–612. - PubMed
-
- Borycki A. G., Li J., Jin F., Emerson C. P., Epstein J. A. (1999). Pax3 functions in cell survival and in pax7 regulation. Dev. Camb. Engl. 126 1665–1674. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

