Liprin-α-Mediated Assemblies and Their Roles in Synapse Formation
- PMID: 33869211
- PMCID: PMC8044993
- DOI: 10.3389/fcell.2021.653381
Liprin-α-Mediated Assemblies and Their Roles in Synapse Formation
Abstract
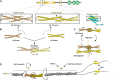
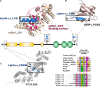
Brain's functions, such as memory and learning, rely on synapses that are highly specialized cellular junctions connecting neurons. Functional synapses orchestrate the assembly of ion channels, receptors, enzymes, and scaffold proteins in both pre- and post-synapse. Liprin-α proteins are master scaffolds in synapses and coordinate various synaptic proteins to assemble large protein complexes. The functions of liprin-αs in synapse formation have been largely uncovered by genetic studies in diverse model systems. Recently, emerging structural and biochemical studies on liprin-α proteins and their binding partners begin to unveil the molecular basis of the synaptic assembly. This review summarizes the recent structural findings on liprin-αs, proposes the assembly mechanism of liprin-α-mediated complexes, and discusses the liprin-α-organized assemblies in the regulation of synapse formation and function.
Keywords: LLPS; SYD2; coiled coil; presynaptic active zone; protein structure; protein–protein interaction; scaffold protein.
Copyright © 2021 Xie, Liang, Yu and Wei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ackley B. D., Harrington R. J., Hudson M. L., Williams L., Kenyon C. J., Chisholm A. D., et al. (2005). The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J. Neurosci. 25 7517–7528. - PMC - PubMed
-
- Arroyo J. D., Lee G. M., Hahn W. C. (2008). Liprin alpha1 interacts with PP2A B56gamma. Cell Cycle 7 525–532. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

