Smyd1 Orchestrates Early Heart Development Through Positive and Negative Gene Regulation
- PMID: 33869215
- PMCID: PMC8047137
- DOI: 10.3389/fcell.2021.654682
Smyd1 Orchestrates Early Heart Development Through Positive and Negative Gene Regulation
Abstract
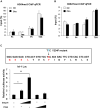
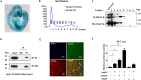
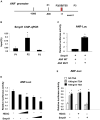
SET and MYND domain-containing protein 1 (Smyd1) is a striated muscle-specific histone methyltransferase. Our previous work demonstrated that deletion of Smyd1 in either cardiomyocytes or the outflow tract (OFT) resulted in embryonic lethality at E9.5, with cardiac structural defects such as truncation of the OFT and right ventricle and impaired expansion and proliferation of the second heart field (SHF). The cardiac phenotype was accompanied by the downregulation of ISL LIM Homeobox 1 (Isl1) and upregulation of atrial natriuretic factor (ANF). However, the mechanisms of Smyd1 regulating Isl1 and ANF during embryonic heart development remain to be elucidated. Here, we employed various biochemical and molecular biological approaches including chromatin immunoprecipitation polymerase chain reaction (ChIP-PCR), pGL3 fluorescence reporter system, and co-immunoprecipitation (CoIP) and found that Smyd1 interacted with absent small homeotic-2-like protein (ASH2L) and activated the promoter of Isl1 by trimethylating H3K4. We also found that Smyd1 associated with HDAC to repress ANF expression using trichostatin A (TSA), a deacetylase inhibitor. In conclusion, Smyd1 participates in early heart development by upregulating the expression of Isl1 and downregulating the expression of ANF.
Keywords: ANF; ASH2L; ISL-1; Smyd1; heart.
Copyright © 2021 Wang, Schwartz, Liu, Sun, Li and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Smyd1 facilitates heart development by antagonizing oxidative and ER stress responses.PLoS One. 2015 Mar 24;10(3):e0121765. doi: 10.1371/journal.pone.0121765. eCollection 2015. PLoS One. 2015. PMID: 25803368 Free PMC article.
-
Defective myogenesis in the absence of the muscle-specific lysine methyltransferase SMYD1.Dev Biol. 2016 Feb 1;410(1):86-97. doi: 10.1016/j.ydbio.2015.12.005. Epub 2015 Dec 11. Dev Biol. 2016. PMID: 26688546
-
Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart.Proc Natl Acad Sci U S A. 2018 Aug 14;115(33):E7871-E7880. doi: 10.1073/pnas.1800680115. Epub 2018 Jul 30. Proc Natl Acad Sci U S A. 2018. PMID: 30061404 Free PMC article.
-
Signaling mechanisms for the activation of an embryonic gene program during the hypertrophy of cardiac ventricular muscle.Basic Res Cardiol. 1992;87 Suppl 2:49-58. doi: 10.1007/978-3-642-72477-0_5. Basic Res Cardiol. 1992. PMID: 1299210 Review.
-
SMYD proteins: key regulators in skeletal and cardiac muscle development and function.Anat Rec (Hoboken). 2014 Sep;297(9):1650-62. doi: 10.1002/ar.22972. Anat Rec (Hoboken). 2014. PMID: 25125178 Review.
Cited by
-
The Methyltransferase Smyd1 Mediates LPS-Triggered Up-Regulation of IL-6 in Endothelial Cells.Cells. 2021 Dec 13;10(12):3515. doi: 10.3390/cells10123515. Cells. 2021. PMID: 34944023 Free PMC article.
-
Autotaxin Inhibition Reduces Post-Ischemic Myocardial Inflammation via Epigenetic Gene Modifications.Stem Cell Rev Rep. 2024 Oct;20(7):1971-1980. doi: 10.1007/s12015-024-10759-7. Epub 2024 Jul 10. Stem Cell Rev Rep. 2024. PMID: 38985374
-
Expression of Smyd1b_tv1 by Alternative Splicing in Cardiac Muscle is Critical for Sarcomere Organization in Cardiomyocytes and Heart Function.Mol Cell Biol. 2024;44(12):543-561. doi: 10.1080/10985549.2024.2402660. Epub 2024 Sep 25. Mol Cell Biol. 2024. PMID: 39320962
-
SMYD1 modulates the proliferation of multipotent cardiac progenitor cells derived from human pluripotent stem cells during myocardial differentiation through GSK3β/β-catenin&ERK signaling.Stem Cell Res Ther. 2024 Oct 8;15(1):350. doi: 10.1186/s13287-024-03899-7. Stem Cell Res Ther. 2024. PMID: 39380045 Free PMC article.
-
Targeting Epigenetic Changes Mediated by Members of the SMYD Family of Lysine Methyltransferases.Molecules. 2023 Feb 20;28(4):2000. doi: 10.3390/molecules28042000. Molecules. 2023. PMID: 36838987 Free PMC article. Review.
References
-
- Berger S. L. (2002). Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12 142–148. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

