Structural Basis of Inhibition of Human Insulin-Regulated Aminopeptidase (IRAP) by Benzopyran-Based Inhibitors
- PMID: 33869280
- PMCID: PMC8047434
- DOI: 10.3389/fmolb.2021.625274
Structural Basis of Inhibition of Human Insulin-Regulated Aminopeptidase (IRAP) by Benzopyran-Based Inhibitors
Abstract
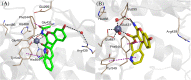
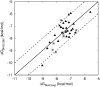
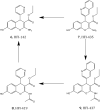
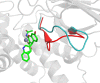
Inhibition of the insulin-regulated aminopeptidase (IRAP) improves memory and cognition in animal models. The enzyme has recently been crystallized and several series of inhibitors reported. We herein focused on one series of benzopyran-based inhibitors of IRAP known as the HFI series, with unresolved binding mode to IRAP, and developed a robust computational model to explain the structure-activity relationship (SAR) and potentially guide their further optimization. The binding model here proposed places the benzopyran ring in the catalytic binding site, coordinating the Zn2+ ion through the oxygen in position 3, in contrast to previous hypothesis. The whole series of HFI compounds was then systematically simulated, starting from this binding mode, using molecular dynamics and binding affinity estimated with the linear interaction energy (LIE) method. The agreement with experimental affinities supports the binding mode proposed, which was further challenged by rigorous free energy perturbation (FEP) calculations. Here, we found excellent correlation between experimental and calculated binding affinity differences, both between selected compound pairs and also for recently reported experimental data concerning the site directed mutagenesis of residue Phe544. The computationally derived structure-activity relationship of the HFI series and the understanding of the involvement of Phe544 in the binding of this scaffold provide valuable information for further lead optimization of novel IRAP inhibitors.
Keywords: Insulin regulated aminopeptidase (IRAP); benzopyran; free energy perturbation (FEP); linear interaction energy (LIE); molecular dynamics (MD).
Copyright © 2021 Vanga, Åqvist, Hallberg and Gutiérrez-de-Terán.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

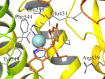

References
-
- Albiston A. L., Pham V., Ye S., Ng L., Lew R. A., Thompson P. E., et al. (2010). Phenylalanine-544 plays a key role in substrate and inhibitor binding by providing a hydrophobic packing point at the active site of insulin-regulated aminopeptidase. Mol. Pharmacol. 78 600–607. 10.1124/mol.110.065458 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

