Structural Analysis of Anti-Hapten Antibodies to Identify Long-Range Structural Movements Induced by Hapten Binding
- PMID: 33869281
- PMCID: PMC8044860
- DOI: 10.3389/fmolb.2021.633526
Structural Analysis of Anti-Hapten Antibodies to Identify Long-Range Structural Movements Induced by Hapten Binding
Abstract
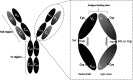
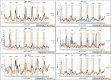
Antibodies are well known for their high specificity that has enabled them to be of significant use in both therapeutic and diagnostic applications. Antibodies can recognize different antigens, including proteins, carbohydrates, peptides, nucleic acids, lipids, and small molecular weight haptens that are abundantly available as hormones, pharmaceuticals, and pesticides. Here we focus on a structural analysis of hapten-antibody couples and identify potential structural movements originating from the hapten binding by comparison with unbound antibody, utilizing 40 crystal structures from the Protein Data Bank. Our analysis reveals three binding surface trends; S1 where a pocket forms to accommodate the hapten, S2 where a pocket is removed when the hapten binds, and S3 where no pockets changes are found. S1 and S2 are expected for induced-fit binding, whereas S3 indicates that a pre-existing population of optimal binding antibody conformation exists. The structural analysis reveals four classifications of structural reorganization, some of which correlate to S2 but not to the other binding surface changes. These observations demonstrate the complexity of the antibody-antigen interaction, where structural changes can be restricted to the binding sites, or extend through the constant domains to propagate structural changes. This highlights the importance of structural analysis to ensure successful and compatible transformation of small antibody fragments at the early discovery stage into full antibodies during the subsequent development stages, where long-range structural changes are required for an Fc effector response.
Keywords: RMSD; RMSF; allosteric movement; antibody; antigen binding fragments (Fab); hapten.
Copyright © 2021 Al Qaraghuli, Kubiak-Ossowska, Ferro and Mulheran.
Conflict of interest statement
Author MQ was employed by the SiMologics Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Structure and specificity of the anti-digoxin antibody 40-50.J Mol Biol. 1995 Apr 28;248(2):344-60. doi: 10.1016/s0022-2836(95)80055-7. J Mol Biol. 1995. PMID: 7739045
-
Structural insights into the evolution of an antibody combining site.Science. 1997 Jun 13;276(5319):1665-9. doi: 10.1126/science.276.5319.1665. Science. 1997. PMID: 9180069
-
A comparative analysis of the immunological evolution of antibody 28B4.Biochemistry. 2001 Sep 11;40(36):10764-73. doi: 10.1021/bi010536c. Biochemistry. 2001. PMID: 11535051
-
Structural aspects of antibodies and antibody-antigen complexes.Ciba Found Symp. 1991;159:13-28; discussion 28-39. doi: 10.1002/9780470514108.ch3. Ciba Found Symp. 1991. PMID: 1959445 Review.
-
Engineered hapten-binding antibody derivatives for modulation of pharmacokinetic properties of small molecules and targeted payload delivery.Immunol Rev. 2016 Mar;270(1):165-77. doi: 10.1111/imr.12386. Immunol Rev. 2016. PMID: 26864111 Free PMC article. Review.
Cited by
-
Drastic alterations in the loop structure around colchicine upon complex formation with an engineered lipocalin indicate a conformational selection mechanism.Acta Crystallogr F Struct Biol Commun. 2023 Sep 1;79(Pt 9):231-239. doi: 10.1107/S2053230X23006817. Epub 2023 Aug 16. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 37584182 Free PMC article.
-
Yeast Surface Display Platform for Rapid Selection of an Antibody Library via Sequential Counter Antigen Flow Cytometry.Antibodies (Basel). 2022 Sep 27;11(4):61. doi: 10.3390/antib11040061. Antibodies (Basel). 2022. PMID: 36278614 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

