High Levels of Antibiotic Resistance in Isolates From Diseased Livestock
- PMID: 33869326
- PMCID: PMC8047425
- DOI: 10.3389/fvets.2021.652351
High Levels of Antibiotic Resistance in Isolates From Diseased Livestock
Abstract
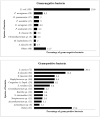
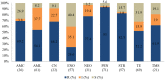
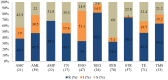
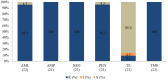
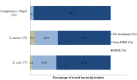
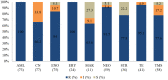
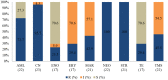
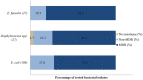
Overuse of antimicrobials in livestock health and production beyond therapeutic needs has been highlighted in recent years as one of the major risk factors for the acceleration of antimicrobial resistance (AMR) of bacteria in both humans and animals. While there is an abundance of reports on AMR in clinical isolates from humans, information regarding the patterns of resistance in clinical isolates from animals is scarce. Hence, a situational analysis of AMR based on clinical isolates from a veterinary diagnostic laboratory was performed to examine the extent and patterns of resistance demonstrated by isolates from diseased food animals. Between 2015 and 2017, 241 cases of diseased livestock were received. Clinical specimens from ruminants (cattle, goats and sheep), and non-ruminants (pigs and chicken) were received for culture and sensitivity testing. A total of 701 isolates were recovered from these specimens. From ruminants, Escherichia coli (n = 77, 19.3%) predominated, followed by Staphylococcus aureus (n = 73, 18.3%). Antibiotic sensitivity testing (AST) revealed that E. coli resistance was highest for penicillin, streptomycin, and neomycin (77-93%). In addition, S. aureus was highly resistant to neomycin, followed by streptomycin and ampicillin (68-82%). More than 67% of E. coli isolates were multi-drug resistant (MDR) and only 2.6% were susceptible to all the tested antibiotics. Similarly, 65.6% of S. aureus isolates were MDR and only 5.5% were susceptible to all tested antibiotics. From non-ruminants, a total of 301 isolates were recovered. Escherichia coli (n = 108, 35.9%) and Staphylococcus spp. (n = 27, 9%) were the most frequent isolates obtained. For E. coli, the highest resistance was against amoxicillin, erythromycin, tetracycline, and neomycin (95-100%). Staphylococcus spp. had a high level of resistance to streptomycin, trimethoprim/sulfamethoxazole, tetracycline and gentamicin (80-100%). The MDR levels of E. coli and Staphylococcus spp. isolates from non-ruminants were 72.2 and 74.1%, respectively. Significantly higher resistance level were observed among isolates from non-ruminants compared to ruminants for tetracycline, amoxicillin, enrofloxacin, and trimethoprim/sulfamethoxazole.
Keywords: Escherichia coli; antimicrobial resistance; cattle; clinical; livestock; pigs.
Copyright © 2021 Haulisah, Hassan, Bejo, Jajere and Ahmad.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- World Organization for Animal Health . OIE List of Antimicrobials of Veterinary Importance (2007). Available online at: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf... (accessed September 12, 2020).
-
- Food Agriculture Organization/World Health Organization/World Organization for Animal Health . Report of the FAO/WHO/OIE Expert Meeting FAO. In: FAO/WHO/OIE Stakeholders Meeting on Critically Important Antimicrobials. (2008). Available online at: http://www.fao.org/3/a-i0204e.pdf (accessed September 12, 2020).
-
- FAAIR Scientific Advisory Panel . A Report of the Facts about Antibiotics in Animals and the Impact on Resistance (FAAIR) Project. The Need to Improve Antimicrobial Use in Agriculture Ecological and Human; (2002). Available online at: https://www.iatp.org/sites/default/files/64_2_36922.pdf (accessed September 13, 2020).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

