LINC00461 facilitates HNSCC development and reduces chemosensitivity by impairing miR-195-mediated inhibition of HOXA10
- PMID: 33869744
- PMCID: PMC8027536
- DOI: 10.1016/j.omto.2021.01.008
LINC00461 facilitates HNSCC development and reduces chemosensitivity by impairing miR-195-mediated inhibition of HOXA10
Abstract
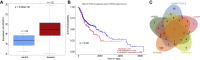
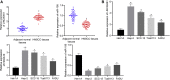
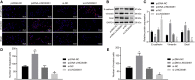
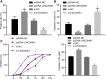
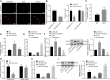
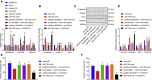
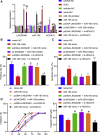
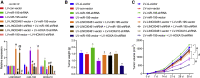
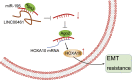
Homeobox A10 (HOXA10) has been regarded to serve as an oncogene in head and neck squamous cell carcinoma (HNSCC). This study was intended to explore the interaction among the long intergenic noncoding RNA 00461 (LINC00461), microRNA (miR)-195, and HOXA10, and to investigate its role in epithelial-mesenchymal transition (EMT) and chemoresistance in HNSCC. The effects of LINC00461, miR-195, and HOXA10 on the EMT and chemoresistance of HNSCC cells were analyzed by comprehensive analysis of gain- and loss-of-function techniques. The intimate relationships among LINC00461, miR-195, and HOXA10 were investigated by several procedures such as RNA-binding protein immunoprecipitation, RNA pull-down, and dual-luciferase reporter assays. A xenotransplantation tumor model in nude mice was established for the assessment of the tumorigenic ability of the cells in vivo. Our findings indicated that LINC00461 was highly expressed in HNSCC and its overexpression induced EMT and precipitated the chemoresistance of HNSCC cells to cisplatin. The LINC00461 could bind to miR-195 while miR-195 targeted HOXA10 independently. Moreover, LINC00461 impaired miR-195-mediated inhibition of HOXA10 to induce EMT and increase the chemoresistance in HNSCC. Tumor weight and volume were reduced by lentivirus-mediated elevation of miR-195 by inhibition of HOXA10, which could be annulled by LINC00461 overexpression. LINC00461 downregulates the expression of miR-195 to subsequently upregulate the expression of HOXA10, thereby promoting EMT and enhancing chemoresistance in HNSCC.
Keywords: chemoresistance; epithelial-mesenchymal transition; head and neck squamous cell carcinoma; homeobox A10; long intergenic noncoding RNA 00461; microRNA-195.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Solomon B., Young R.J., Rischin D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin. Cancer Biol. 2018;52:228–240. - PubMed
-
- Marur S., Forastiere A.A. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2016;91:386–396. - PubMed
-
- Rieke D.T., Klinghammer K., Keilholz U. Targeted therapy of head and neck cancer. Oncol. Res. Treat. 2016;39:780–786. - PubMed
-
- Kaidar-Person O., Gil Z., Billan S. Precision medicine in head and neck cancer. Drug Resist. Updat. 2018;40:13–16. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

