Hepatitis B virus resistance to tenofovir: fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms
- PMID: 33869791
- PMCID: PMC8033640
- DOI: 10.12688/wellcomeopenres.15992.1
Hepatitis B virus resistance to tenofovir: fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms
Abstract
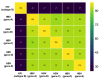
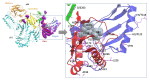
Background: Tenofovir (TFV) is a widely used treatment for chronic hepatitis B virus (HBV) infection. There is a high genetic barrier to the selection of TFV resistance-associated mutations (RAMs), but the distribution and clinical significance of TFV RAMs are not well understood. We here present assimilated evidence for putative TFV RAMs with the aims of cataloguing and characterising mutations that have been reported, and starting to develop insights into mechanisms of resistance. Methods: We carried out a systematic literature search in PubMed and Scopus to identify clinical, in vitro and in silico evidence of TFV resistance. We included peer-reviewed studies presenting original data regarding virological TFV breakthrough, using published methods to assess the quality of each study. We generated a list of RAMs that have been reported in association with TFV resistance, developing a 'long-list' (all reported RAMs) and a 'short-list' (a refined list supported by the most robust evidence). We assessed the potential functional and structural consequences by mapping onto the crystal structure for HIV reverse transcriptase (RT), as the structure of HBV RT has not been solved. Results: We identified a 'long-list' of 37 putative TFV RAMs in HBV RT, occurring within and outside sites of enzyme activity, some of which can be mapped onto a homologous HIV RT structure. A 'short-list' of nine sites are supported by the most robust evidence. If clinically significant resistance arises, it is most likely to be in the context of suites of multiple RAMs. Other factors including adherence, viral load, HBeAg status, HIV coinfection and NA dosage may also influence viraemic suppression. Conclusion: There is emerging evidence for polymorphisms that may reduce susceptibility to TVF. However, good correlation between viral sequence and treatment outcomes is currently lacking; further studies are essential to optimise individual treatment and public health approaches.
Keywords: HBV; Hepatitis B virus; RAMs; TAF; TDF; TFV; Tenofovir; resistance.
Copyright: © 2020 Mokaya J et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


Similar articles
-
Evidence of tenofovir resistance in chronic hepatitis B virus (HBV) infection: An observational case series of South African adults.J Clin Virol. 2020 Aug;129:104548. doi: 10.1016/j.jcv.2020.104548. Epub 2020 Jul 8. J Clin Virol. 2020. PMID: 32663786 Free PMC article.
-
Rare emergence of drug resistance in HIV-1 treatment-naïve patients after 48 weeks of treatment with elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide.HIV Clin Trials. 2016 Mar;17(2):78-87. doi: 10.1080/15284336.2016.1142731. HIV Clin Trials. 2016. PMID: 26892863 Clinical Trial.
-
Anti-hepatitis B virus activity in vitro of combinations of tenofovir with nucleoside/nucleotide analogues.Antivir Chem Chemother. 2009;19(4):165-76. doi: 10.1177/095632020901900404. Antivir Chem Chemother. 2009. PMID: 19374144
-
Antivirals for prevention of hepatitis B virus mother-to-child transmission in human immunodeficiency virus positive pregnant women co-infected with hepatitis B virus.Cochrane Database Syst Rev. 2023 Jun 12;6(6):CD013653. doi: 10.1002/14651858.CD013653.pub2. Cochrane Database Syst Rev. 2023. PMID: 37306558 Free PMC article. Review.
-
The effectiveness of tenofovir-based pre-exposure prophylaxis for prevention of HIV acquisition among sub-Saharan African women at high risk: a systematic review.Pan Afr Med J. 2021 Mar 26;38:308. doi: 10.11604/pamj.2021.38.308.26014. eCollection 2021. Pan Afr Med J. 2021. PMID: 34178226 Free PMC article.
Cited by
-
Viral hepatitis: Milestones, unresolved issues, and future goals.World J Gastroenterol. 2021 Jul 28;27(28):4603-4638. doi: 10.3748/wjg.v27.i28.4603. World J Gastroenterol. 2021. PMID: 34366625 Free PMC article. Review.
-
High Prevalence of Hepatitis B Virus Drug Resistance Mutations to Lamivudine among People with HIV/HBV Coinfection in Rural and Peri-Urban Communities in Botswana.Viruses. 2024 Apr 11;16(4):592. doi: 10.3390/v16040592. Viruses. 2024. PMID: 38675933 Free PMC article.
-
Endemic HBV among hospital in-patients in Bangladesh, including evidence of occult infection.J Gen Virol. 2021 Jul;102(7):001628. doi: 10.1099/jgv.0.001628. J Gen Virol. 2021. PMID: 34328828 Free PMC article.
-
Persistent Hepatitis B Viraemia with Polymerase Mutations among HIV/HBV Co-Infected Patients on HBV-Active ART in KwaZulu-Natal, South Africa.Viruses. 2022 Apr 10;14(4):788. doi: 10.3390/v14040788. Viruses. 2022. PMID: 35458518 Free PMC article.
-
Hepatitis B virus genotype distribution and mutation patterns: Insights and clinical implications for hepatitis B virus positive patients.World J Exp Med. 2025 Jun 20;15(2):102395. doi: 10.5493/wjem.v15.i2.102395. eCollection 2025 Jun 20. World J Exp Med. 2025. PMID: 40546674 Free PMC article. Review.
References
-
- Beloukas A, Geretti AM: Hepatitis B Virus Drug Resistance.In: Antimicrobial Drug Resistance. Cham: Springer International Publishing;2017;1227–42. 10.1007/978-3-319-47266-9_26 - DOI
-
- Mokaya J, Marsden BD, McNaughton A, et al. : Tenofovir resistance in HBV. figshare.Dataset.2020. 10.6084/m9.figshare.8427746.v2 - DOI
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

