Whole genome deep sequencing analysis of viral quasispecies diversity and evolution in HBeAg seroconverters
- PMID: 33870157
- PMCID: PMC8042178
- DOI: 10.1016/j.jhepr.2021.100254
Whole genome deep sequencing analysis of viral quasispecies diversity and evolution in HBeAg seroconverters
Abstract
Background & aims: We aimed to investigate how viral quasispecies of the HBV whole genome evolves and diversifies in response to HBeAg seroconversion and viral control utilising next-generation sequencing (NGS).
Methods: Fifty HBeAg-positive chronic hepatitis B patients, including 18 treatment-naïve and 32 interferon (IFN)-treated individuals, were recruited. Serial HBV whole genomes in serum were analysed by NGS to determine sequence characteristics and viral quasispecies.
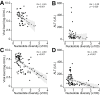
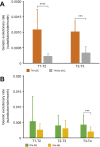
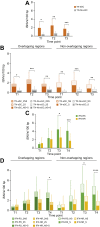
Results: HBV quasispecies diversity, measured by nucleotide diversity, was negatively correlated with viral load and hepatitis activity. Spontaneous HBeAg seroconverters exhibited significantly greater viral quasispecies diversity than treatment-naïve non-seroconverters from >1 year before seroconversion (0.0112 vs. 0.0060, p <0.01) to >1 year after seroconversion (0.0103 vs. 0.0068, p <0.01). IFN-induced HBeAg seroconverters tended to have higher viral genetic diversity than non-seroconverters along with treatment. Particularly, the IFN responders, defined as IFN-induced HBeAg seroconversion with low viraemia, exhibited significantly greater genetic diversity of whole HBV genome at 6 months post-IFN treatment than IFN non-responders (0.0148 vs. 0.0106, p = 0.048). Moreover, spontaneous HBeAg seroconverters and IFN responders exhibited significantly higher evolutionary rates and more intra-host single-nucleotide variants. Interestingly, in spontaneous HBeAg seroconverters and IFN responders, there were distinct evolutionary patterns in the HBV genome.
Conclusions: Higher HBV quasispecies diversity is associated with spontaneous HBeAg seroconversion and IFN-induced HBeAg seroconversion with low viraemia, conferring a favourable clinical outcome.
Lay summary: HBeAg seroconversion is a landmark in the natural history of chronic HBV infection. Using next-generation sequencing, we found that the nucleotide diversity of HBV was negatively correlated with viral load and hepatitis activity. Patients undergoing HBeAg seroconversion had more diverse HBV genomes and a faster viral evolution rate. Our findings suggest HBeAg seroconversion is driven by host selection pressure, likely immune selection pressure.
Keywords: ALT, alanine aminotransferase; AUC, area under curve; BCP, basal core promoter; C, core; CHB, chronic hepatitis B; Chronic hepatitis B; EOT, end of treatment; HBeAg seroconversion; IFN, interferon; IFN-NR, IFN-non-responders; IFN-No-eSC, IFN-treated HBeAg non-seroconverters; IFN-RS, IFN-responders; IFN-eSC, IFN-treated HBeAg seroconverters; Intra-host single nucleotide variants; NGS, next-generation sequencing; ORFs, open reading frames; P, polymerase; S, surface; TN-No-eSC, treatment-naïve non-seroconverters; TN-eSC, treatment-naïve HBeAg seroconverters; dN, nonsynonymous substitution rate; dS, synonymous substitution rate; iSNVs, intra-host single-nucleotide variants.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures





References
-
- Kao J.H. Diagnosis of hepatitis B virus infection through serological and virological markers. Expert Rev Gastroenterol Hepatol. 2008;2:553–562. - PubMed
-
- Yang H.C., Shih Y.F., Liu C.J. Viral factors affecting the clinical outcomes of chronic hepatitis B. J Infect Dis. 2017;216:S757–S764. - PubMed
-
- Nowak M.A., Bangham C.R. Population dynamics of immune responses to persistent viruses. Science. 1996;272:74–79. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

