Severe Acute Respiratory Syndrome Coronavirus 2 Total and Subgenomic RNA Viral Load in Hospitalized Patients
- PMID: 33870434
- PMCID: PMC8083294
- DOI: 10.1093/infdis/jiab215
Severe Acute Respiratory Syndrome Coronavirus 2 Total and Subgenomic RNA Viral Load in Hospitalized Patients
Abstract
Background: Previous studies demonstrated that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA can be detected for weeks after infection. The significance of this finding is unclear and, in most patients, does not represent active infection. Detection of subgenomic RNA has been proposed to represent productive infection and may be a useful marker for monitoring infectivity.
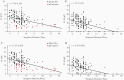
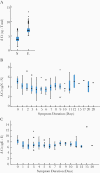
Methods: We used quantitative reverse-transcription polymerase chain reaction (RT-qPCR) to quantify total and subgenomic nucleocapsid (sgN) and envelope (sgE) transcripts in 185 SARS-CoV-2-positive nasopharyngeal swab samples collected on hospital admission and to relate to symptom duration.
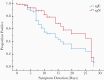
Results: We find that all transcripts decline at the same rate; however, sgE becomes undetectable before other transcripts. The median duration of symptoms to a negative test is 14 days for sgE and 25 days for sgN. There is a linear decline in subgenomic compared to total RNA, suggesting that subgenomic transcript copy number is dependent on copy number of total transcripts. The mean difference between total and sgN is 16-fold and the mean difference between total and sgE is 137-fold. This relationship is constant over duration of symptoms, allowing prediction of subgenomic copy number from total copy number.
Conclusions: Subgenomic RNA may be no more useful in determining infectivity than a copy number threshold determined for total RNA.
Keywords: RT-qPCR; SARS-CoV-2; subgenomic RNA.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures

Update of
-
SARS-CoV-2 Total and Subgenomic RNA Viral Load in Hospitalized Patients.medRxiv [Preprint]. 2021 Mar 1:2021.02.25.21252493. doi: 10.1101/2021.02.25.21252493. medRxiv. 2021. Update in: J Infect Dis. 2021 Oct 28;224(8):1287-1293. doi: 10.1093/infdis/jiab215. PMID: 33688671 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

