The screening of immune-related biomarkers for prognosis of lung adenocarcinoma
- PMID: 33870858
- PMCID: PMC8806236
- DOI: 10.1080/21655979.2021.1911211
The screening of immune-related biomarkers for prognosis of lung adenocarcinoma
Abstract
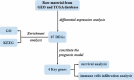
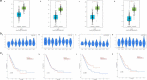
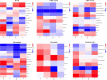
Lung adenocarcinoma (LUAD) accounts for a frequently seen non-small cell lung cancer (NSCLC) histological subtype, and it is associated with dismal prognostic outcome. However, the benefits of traditional treatment are still limited, and the efficacies of immunotherapy are quite different. Therefore, it is of great significance to identify novel immune-related therapeutic targets in lung adenocarcinoma. In this study, we identified a set of immune-related biomarkers for prognosis of lung adenocarcinoma, which could provide new ideas for immunotherapy of lung adenocarcinoma. Datasets related to LUAD were filtered from the GEO database. The appropriate packages were used to identify differentially expressed genes (DEGs) and to carry out enrichment analysis, followed by the construction of prognostic biomarkers. The Kaplan-Meier (K-M) curves were plotted to analyze patient survival based on hub genes. Associations between the expression of selected biomarkers and six types of tumor-infiltrating immune cells were evaluated based on the online tool TIMER. After analyzing five GEO datasets(GSE32867, GSE46539, GSE63459, GSE75037 and GSE116959), we discovered altogether 67 DEGs, among which, 15 showed up-regulation while 52 showed down-regulation. Enrichments of integrated DEGs were identified in the ontology categories. CAV1, CFD, FMO2 and CLEC3B were eventually selected as independent prognostic biomarkers, they were correlated with clinical outcomes of LUAD patients. Moreover, a positive correlation was observed between biomarker expression and all different types of immune infiltration, and the expression level of the four biomarkers was all positively related to macrophage.
Keywords: Lung adenocarcinoma (LUAD); bioinformatics analysis; immune infiltration.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Bc B, Dela Cruz CS.. Lung Cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24. - PubMed
-
- Torre LA, Siegel RL, Jemal A.. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. - PubMed
-
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
