MAUI (MBI Analysis User Interface)-An image processing pipeline for Multiplexed Mass Based Imaging
- PMID: 33872301
- PMCID: PMC8084329
- DOI: 10.1371/journal.pcbi.1008887
MAUI (MBI Analysis User Interface)-An image processing pipeline for Multiplexed Mass Based Imaging
Abstract
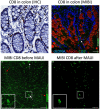
Mass Based Imaging (MBI) technologies such as Multiplexed Ion Beam Imaging by time of flight (MIBI-TOF) and Imaging Mass Cytometry (IMC) allow for the simultaneous measurement of the expression levels of 40 or more proteins in biological tissue, providing insight into cellular phenotypes and organization in situ. Imaging artifacts, resulting from the sample, assay or instrumentation complicate downstream analyses and require correction by domain experts. Here, we present MBI Analysis User Interface (MAUI), a series of graphical user interfaces that facilitate this data pre-processing, including the removal of channel crosstalk, noise and antibody aggregates. Our software streamlines these steps and accelerates processing by enabling real-time and interactive parameter tuning across multiple images.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: MA is a consultant and shareholder in IonPath.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

