A unique view of SARS-CoV-2 through the lens of ORF8 protein
- PMID: 33872970
- PMCID: PMC8049180
- DOI: 10.1016/j.compbiomed.2021.104380
A unique view of SARS-CoV-2 through the lens of ORF8 protein
Abstract
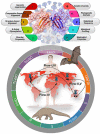
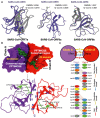
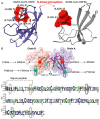
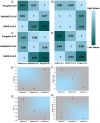
Immune evasion is one of the unique characteristics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) attributed to its ORF8 protein. This protein modulates the adaptive host immunity through down-regulation of MHC-1 (Major Histocompatibility Complex) molecules and innate immune responses by surpassing the host's interferon-mediated antiviral response. To understand the host's immune perspective in reference to the ORF8 protein, a comprehensive study of the ORF8 protein and mutations possessed by it have been performed. Chemical and structural properties of ORF8 proteins from different hosts, such as human, bat, and pangolin, suggest that the ORF8 of SARS-CoV-2 is much closer to ORF8 of Bat RaTG13-CoV than to that of Pangolin-CoV. Eighty-seven mutations across unique variants of ORF8 in SARS-CoV-2 can be grouped into four classes based on their predicted effects (Hussain et al., 2021) [1]. Based on the geo-locations and timescale of sample collection, a possible flow of mutations was built. Furthermore, conclusive flows of amalgamation of mutations were found upon sequence similarity analyses and consideration of the amino acid conservation phylogenies. Therefore, this study seeks to highlight the uniqueness of the rapidly evolving SARS-CoV-2 through the ORF8.
Keywords: Mutational hotspots; ORF8; ORF8 evolution; Phylogenetics; Physicochemical properties; SARS-CoV-2.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
All authors declare no conflict of interest.
Figures



References
-
- Kraemer M.U.G., Yang C.H., Gutierrez B., Wu C.H., Klein B., Pigott D.M., Open C.-D.W.G., du Plessis L., Faria N.R., Li R., Hanage W.P., Brownstein J.S., Layan M., Vespignani A., Tian H., Dye C., Pybus O.G., Scarpino S.V. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science. 2020;368:493–497. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

