Structural Insights into the Mechanisms of Action of Functionally Distinct Classes of Chikungunya Virus Nonstructural Protein 1 Inhibitors
- PMID: 33875421
- PMCID: PMC8218635
- DOI: 10.1128/AAC.02566-20
Structural Insights into the Mechanisms of Action of Functionally Distinct Classes of Chikungunya Virus Nonstructural Protein 1 Inhibitors
Abstract
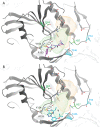
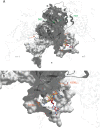
Chikungunya virus (CHIKV) nonstructural protein 1 (nsP1) harbors the methyltransferase (MTase) and guanylyltransferase (GTase) activities needed for viral RNA capping and represents a promising antiviral drug target. We compared the antiviral efficacies of nsP1 inhibitors belonging to the MADTP, CHVB, and FHNA series (6'-fluoro-homoneplanocin A [FHNA], its 3'-keto form, and 6'-β-fluoro-homoaristeromycin). Cell-based phenotypic cross-resistance assays revealed that the CHVB and MADTP series had similar modes of action that differed from that of the FHNA series. In biochemical assays with purified Semliki Forest virus and CHIKV nsP1, CHVB compounds strongly inhibited MTase and GTase activities, while MADTP-372 had a moderate inhibitory effect. FHNA did not directly inhibit the enzymatic activity of CHIKV nsP1. The first-of-their-kind molecular-docking studies with the cryo-electron microscopy (cryo-EM) structure of CHIKV nsP1, which is assembled into a dodecameric ring, revealed that the MADTP and CHVB series bind at the S-adenosylmethionine (SAM)-binding site in the capping domain, where they would function as competitive or noncompetitive inhibitors. The FHNA series was predicted to bind at the secondary binding pocket in the ring-aperture membrane-binding and oligomerization (RAMBO) domain, potentially interfering with the membrane binding and oligomerization of nsP1. Our cell-based and enzymatic assays, in combination with molecular docking and mapping of compound resistance mutations to the nsP1 structure, allowed us to group nsP1 inhibitors into functionally distinct classes. This study identified druggable pockets in the nsP1 dodecameric structure and provides a basis for the rational design, optimization, and combination of inhibitors of this unique and promising antiviral drug target.
Keywords: Chikungunya virus; GTP; SAM; antivirals; binding pocket; capping; inhibitors; molecular docking; nsP1; resistance.
Figures






Similar articles
-
Advances in the Development of Non-Structural Protein 1 (NsP1) Inhibitors for the Treatment of Chikungunya Virus Infection.Mini Rev Med Chem. 2024;24(22):1972-1982. doi: 10.2174/0113895575301735240607055839. Mini Rev Med Chem. 2024. PMID: 38910486 Review.
-
6'-β-Fluoro-Homoaristeromycin and 6'-Fluoro-Homoneplanocin A Are Potent Inhibitors of Chikungunya Virus Replication through Their Direct Effect on Viral Nonstructural Protein 1.Antimicrob Agents Chemother. 2020 Mar 24;64(4):e02532-19. doi: 10.1128/AAC.02532-19. Print 2020 Mar 24. Antimicrob Agents Chemother. 2020. PMID: 31964798 Free PMC article.
-
Novel Class of Chikungunya Virus Small Molecule Inhibitors That Targets the Viral Capping Machinery.Antimicrob Agents Chemother. 2020 Jun 23;64(7):e00649-20. doi: 10.1128/AAC.00649-20. Print 2020 Jun 23. Antimicrob Agents Chemother. 2020. PMID: 32340991 Free PMC article.
-
Identification of small molecule inhibitors of the Chikungunya virus nsP1 RNA capping enzyme.Antiviral Res. 2018 Jun;154:124-131. doi: 10.1016/j.antiviral.2018.03.013. Epub 2018 Apr 20. Antiviral Res. 2018. PMID: 29680670 Free PMC article.
-
Small-Molecule Inhibitors of Chikungunya Virus: Mechanisms of Action and Antiviral Drug Resistance.Antimicrob Agents Chemother. 2020 Nov 17;64(12):e01788-20. doi: 10.1128/AAC.01788-20. Print 2020 Nov 17. Antimicrob Agents Chemother. 2020. PMID: 32928738 Free PMC article. Review.
Cited by
-
Independent repeated mutations within the alphaviruses Ross River virus and Barmah Forest virus indicates convergent evolution and past positive selection in ancestral populations despite ongoing purifying selection.Virus Evol. 2024 Sep 13;10(1):veae080. doi: 10.1093/ve/veae080. eCollection 2024. Virus Evol. 2024. PMID: 39411152 Free PMC article.
-
Advances in antiviral strategies targeting mosquito-borne viruses: cellular, viral, and immune-related approaches.Virol J. 2025 Feb 4;22(1):26. doi: 10.1186/s12985-025-02622-z. Virol J. 2025. PMID: 39905499 Free PMC article. Review.
-
Chemical biology and medicinal chemistry of RNA methyltransferases.Nucleic Acids Res. 2022 May 6;50(8):4216-4245. doi: 10.1093/nar/gkac224. Nucleic Acids Res. 2022. PMID: 35412633 Free PMC article.
-
Chlorinated biscoumarins inhibit chikungunya virus replication in cell-based and animal models.Emerg Microbes Infect. 2025 Dec;14(1):2529889. doi: 10.1080/22221751.2025.2529889. Epub 2025 Jul 28. Emerg Microbes Infect. 2025. PMID: 40608982 Free PMC article.
-
Advances in the Development of Non-Structural Protein 1 (NsP1) Inhibitors for the Treatment of Chikungunya Virus Infection.Mini Rev Med Chem. 2024;24(22):1972-1982. doi: 10.2174/0113895575301735240607055839. Mini Rev Med Chem. 2024. PMID: 38910486 Review.
References
-
- Burt FJ, Chen W, Miner JJ, Lenschow DJ, Merits A, Schnettler E, Kohl A, Rudd PA, Taylor A, Herrero LJ, Zaid A, Ng LFP, Mahalingam S. 2017. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect Dis 17:e107–e117. 10.1016/S1473-3099(16)30385-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

