Pharmacogene Sequencing of a Gabonese Population with Severe Plasmodium falciparum Malaria Reveals Multiple Novel Variants with Putative Relevance for Antimalarial Treatment
- PMID: 33875422
- PMCID: PMC8218688
- DOI: 10.1128/AAC.00275-21
Pharmacogene Sequencing of a Gabonese Population with Severe Plasmodium falciparum Malaria Reveals Multiple Novel Variants with Putative Relevance for Antimalarial Treatment
Abstract
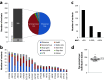
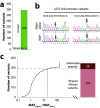
Malaria remains one of the deadliest diseases in Africa, particularly for children. While successful in reducing morbidity and mortality, antimalarial treatments are also a major cause of adverse drug reactions (ADRs). Host genetic variation in genes involved in drug disposition or toxicity constitutes an important determinant of ADR risk and can prime for parasite drug resistance. Importantly, however, the genetic diversity in Africa is substantial, and thus, genetic profiles in one population cannot be reliably extrapolated to other ethnogeographic groups. Gabon is considered a high-transmission country, with more than 460,000 malaria cases per year. Yet the pharmacogenetic landscape of the Gabonese population or its neighboring countries has not been analyzed. Using targeted sequencing, here, we profiled 21 pharmacogenes with importance for antimalarial treatment in 48 Gabonese pediatric patients with severe Plasmodium falciparum malaria. Overall, we identified 347 genetic variants, of which 18 were novel, and each individual was found to carry 87.3 ± 9.2 (standard deviation [SD]) variants across all analyzed genes. Importantly, 16.7% of these variants were population specific, highlighting the need for high-resolution pharmacogenomic profiling. Between one in three and one in six individuals harbored reduced-activity alleles of CYP2A6, CYP2B6, CYP2D6, and CYP2C8 with important implications for artemisinin, chloroquine, and amodiaquine therapy. Furthermore, one in three patients harbored at least one G6PD-deficient allele, suggesting a considerably increased risk of hemolytic anemia upon exposure to aminoquinolines. Combined, our results reveal the unique genetic landscape of the Gabonese population and pinpoint the genetic basis for interindividual differences in antimalarial drug responses and toxicity.
Keywords: Gabon; malaria; population pharmacogenetics; precision medicine; public health.
Figures

References
-
- World Health Organization. 2019. WHO malaria report 2019. World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

