YAF2-Mediated YY1-Sirtuin6 Interactions Responsible for Mitochondrial Downregulation in Aging Tunicates
- PMID: 33875574
- PMCID: PMC8224230
- DOI: 10.1128/MCB.00047-21
YAF2-Mediated YY1-Sirtuin6 Interactions Responsible for Mitochondrial Downregulation in Aging Tunicates
Abstract
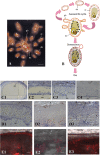
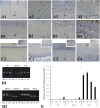
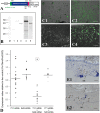
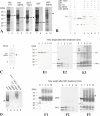
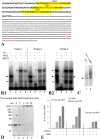
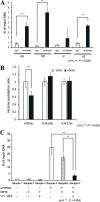
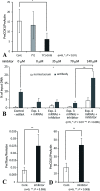
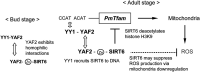
In budding tunicates, aging accompanies a decrease in the gene expression of mitochondrial transcription factor A (Tfam), and the in vivo transfection of Tfam mRNA stimulates the mitochondrial respiratory activity of aged animals. The gene expression of both the transcriptional repressor Yin-Yang-1 (YY1) and corepressor Sirtuin6 (Sirt6) increased during aging, and the cotransfection of synthetic mRNA of YY1 and Sirt6 synergistically downregulated Tfam gene expression. Pulldown assays of proteins indicated that YY1-associated factor 2 (YAF2) was associated with both YY1 and SIRT6. Protein cross-linking confirmed that YAF2 bound YY1 and SIRT6 with a molar ratio of 1:1. YY1 was bound to CCAT- or ACAT-containing oligonucleotides in the 5' flanking region of the Tfam gene. Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) showed that SIRT6 specifically induced the histone H3 lysine 9 (H3K9) deacetylation of the Tfam upstream region. YY1 and YAF2 accelerated SIRT6-induced H3K9 deacetylation. YY1 and Sirt6 mRNA transfection attenuated mitochondrial respiratory gene expression and blocked MitoTracker fluorescence. In contrast, the SIRT6 inhibitor and Tfam mRNA antagonized the inhibitory effects of YY1 and Sirt6, indicating that Tfam acts on mitochondria downstream of YY1 and Sirt6. We concluded that in the budding tunicate Polyandrocarpa misakiensis, YY1 recruits SIRT6 via YAF2 to the TFAM gene, resulting in aging-related mitochondrial downregulation.
Keywords: Tfam; ascidian; budding; epigenetics; gel shift assay; senescence.
Figures

Similar articles
-
Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1.Nucleic Acids Res. 1997 Feb 15;25(4):843-9. doi: 10.1093/nar/25.4.843. Nucleic Acids Res. 1997. PMID: 9016636 Free PMC article.
-
Cooperation between Myc and YY1 provides novel silencing transcriptional targets of alpha3beta1-integrin in tumour cells.Oncogene. 2007 Jan 18;26(3):382-94. doi: 10.1038/sj.onc.1209804. Epub 2006 Jul 31. Oncogene. 2007. PMID: 16878156
-
YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment.Nucleic Acids Res. 2014 Feb;42(4):2208-23. doi: 10.1093/nar/gkt1187. Epub 2013 Nov 26. Nucleic Acids Res. 2014. PMID: 24285299 Free PMC article.
-
Regulation of mitochondrial biogenesis through TFAM-mitochondrial DNA interactions: Useful insights from aging and calorie restriction studies.Mitochondrion. 2015 Nov;25:67-75. doi: 10.1016/j.mito.2015.10.001. Epub 2015 Oct 3. Mitochondrion. 2015. PMID: 26437364 Review.
-
The Yin and Yang of YY1 in tumor growth and suppression.Int J Cancer. 2018 Aug 1;143(3):460-465. doi: 10.1002/ijc.31255. Epub 2018 Jan 31. Int J Cancer. 2018. PMID: 29322514 Review.
Cited by
-
The regulatory network of potential transcription factors and MiRNAs of mitochondria-related genes for sarcopenia.Front Genet. 2022 Sep 12;13:975886. doi: 10.3389/fgene.2022.975886. eCollection 2022. Front Genet. 2022. PMID: 36171891 Free PMC article.
-
Targeting Cellular Senescence in Aging and Age-Related Diseases: Challenges, Considerations, and the Emerging Role of Senolytic and Senomorphic Therapies.Aging Dis. 2024 Feb 27;15(6):2554-2594. doi: 10.14336/AD.2024.0206. Aging Dis. 2024. PMID: 38421832 Free PMC article. Review.
-
LncRNA NORAD Promotes Vascular Endothelial Cell Injury and Atherosclerosis Through Suppressing VEGF Gene Transcription via Enhancing H3K9 Deacetylation by Recruiting HDAC6.Front Cell Dev Biol. 2021 Jul 9;9:701628. doi: 10.3389/fcell.2021.701628. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34307380 Free PMC article.
-
SIRT6 Through the Brain Evolution, Development, and Aging.Front Aging Neurosci. 2021 Oct 13;13:747989. doi: 10.3389/fnagi.2021.747989. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34720996 Free PMC article.
-
DNA stimulates the deacetylase SIRT6 to mono-ADP-ribosylate proteins with histidine repeats.J Biol Chem. 2025 Jun;301(6):108532. doi: 10.1016/j.jbc.2025.108532. Epub 2025 Apr 23. J Biol Chem. 2025. PMID: 40280420 Free PMC article.
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2014. Molecular biology of the cell, 6th ed. Garland Science Taylor and Francis Group, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
