Transcriptomic analysis to identify genes associated with selective hippocampal vulnerability in Alzheimer's disease
- PMID: 33875655
- PMCID: PMC8055900
- DOI: 10.1038/s41467-021-22399-3
Transcriptomic analysis to identify genes associated with selective hippocampal vulnerability in Alzheimer's disease
Abstract
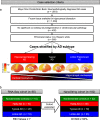
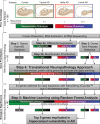
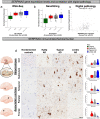
Selective vulnerability of different brain regions is seen in many neurodegenerative disorders. The hippocampus and cortex are selectively vulnerable in Alzheimer's disease (AD), however the degree of involvement of the different brain regions differs among patients. We classified corticolimbic patterns of neurofibrillary tangles in postmortem tissue to capture extreme and representative phenotypes. We combined bulk RNA sequencing with digital pathology to examine hippocampal vulnerability in AD. We identified hippocampal gene expression changes associated with hippocampal vulnerability and used machine learning to identify genes that were associated with AD neuropathology, including SERPINA5, RYBP, SLC38A2, FEM1B, and PYDC1. Further histologic and biochemical analyses suggested SERPINA5 expression is associated with tau expression in the brain. Our study highlights the importance of embracing heterogeneity of the human brain in disease to identify disease-relevant gene expression.
Conflict of interest statement
R.C.P. is a consultant for Hoffman-La Roche, Merck, Biogen, Eisai. R.C.P. is on the Data safety and monitoring board for Genentech. N.G.R. takes part in multi-center studies funded by Lilly, Biogen, and Abbvie. M.E.M. served as a consultant for AVID Radiopharmaceuticals. All other authors declare no competing interests.
Figures



Similar articles
-
The SERPINA5 coding variant E228Q does not contribute to clinicopathologic characteristics in Alzheimer's disease: A cross-sectional study.Medicine (Baltimore). 2023 Jun 16;102(24):e34017. doi: 10.1097/MD.0000000000034017. Medicine (Baltimore). 2023. PMID: 37327267 Free PMC article.
-
A novel transgenic mouse expressing double mutant tau driven by its natural promoter exhibits tauopathy characteristics.Exp Neurol. 2008 Jul;212(1):71-84. doi: 10.1016/j.expneurol.2008.03.007. Epub 2008 Mar 21. Exp Neurol. 2008. PMID: 18490011
-
Molecular properties underlying regional vulnerability to Alzheimer's disease pathology.Brain. 2018 Sep 1;141(9):2755-2771. doi: 10.1093/brain/awy189. Brain. 2018. PMID: 30016411 Free PMC article.
-
[Molecular mechanisms in Alzheimer's disease].Rev Neurol. 1997 Jul;25 Suppl 1:S50-4. Rev Neurol. 1997. PMID: 9280667 Review. Spanish. No abstract available.
-
Regulatable transgenic mouse models of Alzheimer disease: onset, reversibility and spreading of Tau pathology.FEBS J. 2013 Sep;280(18):4371-81. doi: 10.1111/febs.12250. Epub 2013 Apr 22. FEBS J. 2013. PMID: 23517246 Review.
Cited by
-
Impairment of entorhinal cortex network activity in Alzheimer's disease.Front Aging Neurosci. 2024 May 31;16:1402573. doi: 10.3389/fnagi.2024.1402573. eCollection 2024. Front Aging Neurosci. 2024. PMID: 38882526 Free PMC article. Review.
-
A Holistic Analysis of Alzheimer's Disease-Associated lncRNA Communities Reveals Enhanced lncRNA-miRNA-RBP Regulatory Triad Formation Within Functionally Segregated Clusters.J Mol Neurosci. 2024 Aug 15;74(3):77. doi: 10.1007/s12031-024-02244-0. J Mol Neurosci. 2024. PMID: 39143264 Free PMC article.
-
Role of Epigenetic Modulation in Neurodegenerative Diseases: Implications of Phytochemical Interventions.Antioxidants (Basel). 2024 May 15;13(5):606. doi: 10.3390/antiox13050606. Antioxidants (Basel). 2024. PMID: 38790711 Free PMC article. Review.
-
Unraveling Psychiatric Disorders through Neural Single-Cell Transcriptomics Approaches.Genes (Basel). 2023 Mar 22;14(3):771. doi: 10.3390/genes14030771. Genes (Basel). 2023. PMID: 36981041 Free PMC article. Review.
-
Modulation of neural activity and gene expression by arecoline.Front Integr Neurosci. 2025 Apr 9;19:1545260. doi: 10.3389/fnint.2025.1545260. eCollection 2025. Front Integr Neurosci. 2025. PMID: 40271198 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG034676/AG/NIA NIH HHS/United States
- U01 AG057195/AG/NIA NIH HHS/United States
- R01 AG054449/AG/NIA NIH HHS/United States
- RF1 AG051504/AG/NIA NIH HHS/United States
- P50 AG047266/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

