STING enhances cell death through regulation of reactive oxygen species and DNA damage
- PMID: 33875663
- PMCID: PMC8055995
- DOI: 10.1038/s41467-021-22572-8
STING enhances cell death through regulation of reactive oxygen species and DNA damage
Abstract
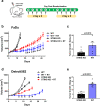
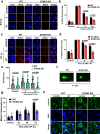
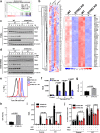
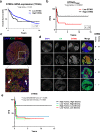
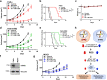
Resistance to DNA-damaging agents is a significant cause of treatment failure and poor outcomes in oncology. To identify unrecognized regulators of cell survival we performed a whole-genome CRISPR-Cas9 screen using treatment with ionizing radiation as a selective pressure, and identified STING (stimulator of interferon genes) as an intrinsic regulator of tumor cell survival. We show that STING regulates a transcriptional program that controls the generation of reactive oxygen species (ROS), and that STING loss alters ROS homeostasis to reduce DNA damage and to cause therapeutic resistance. In agreement with these data, analysis of tumors from head and neck squamous cell carcinoma patient specimens show that low STING expression is associated with worse outcomes. We also demonstrate that pharmacologic activation of STING enhances the effects of ionizing radiation in vivo, providing a rationale for therapeutic combinations of STING agonists and DNA-damaging agents. These results highlight a role for STING that is beyond its canonical function in cyclic dinucleotide and DNA damage sensing, and identify STING as a regulator of cellular ROS homeostasis and tumor cell susceptibility to reactive oxygen dependent, DNA damaging agents.
Conflict of interest statement
J.N.C. has a research agreement with Spring Bank Pharmaceuticals. D.L.R. has served as a consultant, advisor, or served on a Scientific Advisory Board for Amgen, Astra Zeneca, Agendia, Biocept, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, GSK, Merck, NanoString, Perkin Elmer, PAIGE, Sanofi, and Ultivue. He has received research funding from Astra Zeneca, Cepheid, Nanostring, Navigate/Novartis, NextCure, Lilly, Ultivue, and Perkin Elmer. B.A.B. received an honorarium from Aduro. K.L., R.I., and S.C. were employed by Spring Bank Pharmaceuticals/F-STAR. All remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

