On the origin of the widespread self-compatible allotetraploid Capsella bursa-pastoris (Brassicaceae)
- PMID: 33875831
- PMCID: PMC8249383
- DOI: 10.1038/s41437-021-00434-9
On the origin of the widespread self-compatible allotetraploid Capsella bursa-pastoris (Brassicaceae)
Abstract
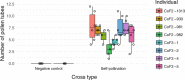
Polyploidy, or whole-genome duplication, is a common speciation mechanism in plants. An important barrier to polyploid establishment is a lack of compatible mates. Because self-compatibility alleviates this problem, it has long been hypothesized that there should be an association between polyploidy and self-compatibility (SC), but empirical support for this prediction is mixed. Here, we investigate whether the molecular makeup of the Brassicaceae self-incompatibility (SI) system, and specifically dominance relationships among S-haplotypes mediated by small RNAs, could facilitate loss of SI in allopolyploid crucifers. We focus on the allotetraploid species Capsella bursa-pastoris, which formed ~300 kya by hybridization and whole-genome duplication involving progenitors from the lineages of Capsella orientalis and Capsella grandiflora. We conduct targeted long-read sequencing to assemble and analyze eight full-length S-locus haplotypes, representing both homeologous subgenomes of C. bursa-pastoris. We further analyze small RNA (sRNA) sequencing data from flower buds to identify candidate dominance modifiers. We find that C. orientalis-derived S-haplotypes of C. bursa-pastoris harbor truncated versions of the male SI specificity gene SCR and express a conserved sRNA-based candidate dominance modifier with a target in the C. grandiflora-derived S-haplotype. These results suggest that pollen-level dominance may have facilitated loss of SI in C. bursa-pastoris. Finally, we demonstrate that spontaneous somatic tetraploidization after a wide cross between C. orientalis and C. grandiflora can result in production of self-compatible tetraploid offspring. We discuss the implications of this finding on the mode of formation of this widespread weed.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bachmann JA, Tedder A, Laenen B, Fracassetti M, Désamoré A, Lafon-Placette C, Steige KA, Callot C, Marande W, Neuffer B, Bergès H, Köhler C, Castric V, Slotte T. Genetic basis and timing of a major mating system shift in Capsella. N. Phytol. 2019;224:505–517. - PubMed
-
- Barrington BC. Polyploidy and self-fertilization in flowering plants. Am J Bot. 2007;94:1527–1533. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

