Large-scale sequencing of flatfish genomes provides insights into the polyphyletic origin of their specialized body plan
- PMID: 33875864
- PMCID: PMC8110480
- DOI: 10.1038/s41588-021-00836-9
Large-scale sequencing of flatfish genomes provides insights into the polyphyletic origin of their specialized body plan
Abstract
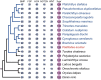
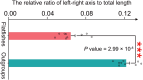
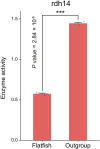
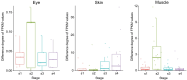
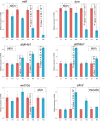
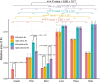
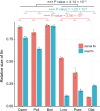
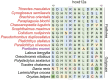
The evolutionary and genetic origins of the specialized body plan of flatfish are largely unclear. We analyzed the genomes of 11 flatfish species representing 9 of the 14 Pleuronectiforme families and conclude that Pleuronectoidei and Psettodoidei do not form a monophyletic group, suggesting independent origins from different percoid ancestors. Genomic and transcriptomic data indicate that genes related to WNT and retinoic acid pathways, hampered musculature and reduced lipids might have functioned in the evolution of the specialized body plan of Pleuronectoidei. Evolution of Psettodoidei involved similar but not identical genes. Our work provides valuable resources and insights for understanding the genetic origins of the unusual body plan of flatfishes.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Phylogenomic analysis of carangimorph fishes reveals flatfish asymmetry arose in a blink of the evolutionary eye.BMC Evol Biol. 2016 Oct 21;16(1):224. doi: 10.1186/s12862-016-0786-x. BMC Evol Biol. 2016. PMID: 27769164 Free PMC article.
-
Mitochondrial genomic investigation of flatfish monophyly.Gene. 2014 Nov 10;551(2):176-82. doi: 10.1016/j.gene.2014.08.053. Epub 2014 Aug 27. Gene. 2014. PMID: 25172210
-
Are flatfishes (Pleuronectiformes) monophyletic?Mol Phylogenet Evol. 2013 Dec;69(3):664-73. doi: 10.1016/j.ympev.2013.07.011. Epub 2013 Jul 19. Mol Phylogenet Evol. 2013. PMID: 23876291 Free PMC article.
-
Flatfish: an asymmetric perspective on metamorphosis.Curr Top Dev Biol. 2013;103:167-94. doi: 10.1016/B978-0-12-385979-2.00006-X. Curr Top Dev Biol. 2013. PMID: 23347519 Review.
-
Genomic resources for flatfish research and their applications.J Fish Biol. 2010 Oct;77(5):1045-70. doi: 10.1111/j.1095-8649.2010.02695.x. J Fish Biol. 2010. PMID: 21039490 Review.
Cited by
-
Reply to: Phylogenomic and comparative genomic analyses support a single evolutionary origin of flatfish asymmetry.Nat Genet. 2024 Jun;56(6):1073-1074. doi: 10.1038/s41588-024-01783-x. Epub 2024 May 27. Nat Genet. 2024. PMID: 38802565 No abstract available.
-
A review of the pangenome: how it affects our understanding of genomic variation, selection and breeding in domestic animals?J Anim Sci Biotechnol. 2023 May 5;14(1):73. doi: 10.1186/s40104-023-00860-1. J Anim Sci Biotechnol. 2023. PMID: 37143156 Free PMC article. Review.
-
Diverse Origins of Near-Identical Antifreeze Proteins in Unrelated Fish Lineages Provide Insights Into Evolutionary Mechanisms of New Gene Birth and Protein Sequence Convergence.Mol Biol Evol. 2024 Sep 4;41(9):msae182. doi: 10.1093/molbev/msae182. Mol Biol Evol. 2024. PMID: 39213383 Free PMC article.
-
Chromosome-level genome assembly and annotation of eel goby (Odontamblyopus rebecca).Sci Data. 2024 Feb 2;11(1):160. doi: 10.1038/s41597-024-02997-8. Sci Data. 2024. PMID: 38307872 Free PMC article.
-
Overview and New Insights Into the Diversity, Evolution, Role, and Regulation of Kisspeptins and Their Receptors in Teleost Fish.Front Endocrinol (Lausanne). 2022 Mar 22;13:862614. doi: 10.3389/fendo.2022.862614. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35392133 Free PMC article. Review.
References
-
- Dornbos SQ, Bottjer DJ, Chen JY. Paleoecology of benthic metazoans in the early cambrian maotianshan shale biota and the middle cambrian burgess shale biota: evidence for the Cambrian substrate revolution. Palaeogeogr. Palaeoclmatol. Palaeoecol. 2005;220:47–67. doi: 10.1016/j.palaeo.2003.11.016. - DOI
-
- Bottjer DJ. The Cambrian substrate revolution and early evolution of the phyla. J. Earth Sci. 2010;21:21–24. doi: 10.1007/s12583-010-0160-7. - DOI
-
- Ryer CH. A review of flatfish behavior relative to trawls. Fish. Res. 2008;90:138–146. doi: 10.1016/j.fishres.2007.10.005. - DOI
-
- Holmes RA, Gibson RN. A comparison of predatory behavior in flatfish. Anim. Behav. 1983;31:1244–1255. doi: 10.1016/S0003-3472(83)80031-1. - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

