An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank
- PMID: 33875891
- PMCID: PMC7610742
- DOI: 10.1038/s41593-021-00826-4
An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank
Abstract
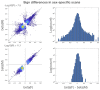
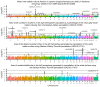
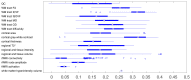
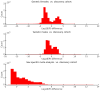
UK Biobank is a major prospective epidemiological study, including multimodal brain imaging, genetics and ongoing health outcomes. Previously, we published genome-wide associations of 3,144 brain imaging-derived phenotypes, with a discovery sample of 8,428 individuals. Here we present a new open resource of genome-wide association study summary statistics, using the 2020 data release, almost tripling the discovery sample size. We now include the X chromosome and new classes of imaging-derived phenotypes (subcortical volumes and tissue contrast). Previously, we found 148 replicated clusters of associations between genetic variants and imaging phenotypes; in this study, we found 692, including 12 on the X chromosome. We describe some of the newly found associations, focusing on the X chromosome and autosomal associations involving the new classes of imaging-derived phenotypes. Our novel associations implicate, for example, pathways involved in the rare X-linked STAR (syndactyly, telecanthus and anogenital and renal malformations) syndrome, Alzheimer's disease and mitochondrial disorders.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
From base pair to brain.Nat Neurosci. 2021 May;24(5):619-621. doi: 10.1038/s41593-021-00852-2. Nat Neurosci. 2021. PMID: 33875895 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

