14q32 rearrangements deregulating BCL11B mark a distinct subgroup of T-lymphoid and myeloid immature acute leukemia
- PMID: 33876209
- PMCID: PMC8513670
- DOI: 10.1182/blood.2020010510
14q32 rearrangements deregulating BCL11B mark a distinct subgroup of T-lymphoid and myeloid immature acute leukemia
Abstract
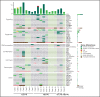
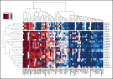
Acute leukemias (ALs) of ambiguous lineage are a heterogeneous group of high-risk leukemias characterized by coexpression of myeloid and lymphoid markers. In this study, we identified a distinct subgroup of immature acute leukemias characterized by a broadly variable phenotype, covering acute myeloid leukemia (AML, M0 or M1), T/myeloid mixed-phenotype acute leukemia (T/M MPAL), and early T-cell precursor acute lymphoblastic leukemia (ETP-ALL). Rearrangements at 14q32/BCL11B are the cytogenetic hallmark of this entity. In our screening of 915 hematological malignancies, there were 202 AML and 333 T-cell acute lymphoblastic leukemias (T-ALL: 58, ETP; 178, non-ETP; 8, T/M MPAL; 89, not otherwise specified). We identified 20 cases of immature leukemias (4% of AML and 3.6% of T-ALL), harboring 4 types of 14q32/BCL11B translocations: t(2,14)(q22.3;q32) (n = 7), t(6;14)(q25.3;q32) (n = 9), t(7;14)(q21.2;q32) (n = 2), and t(8;14)(q24.2;q32) (n = 2). The t(2;14) produced a ZEB2-BCL11B fusion transcript, whereas the other 3 rearrangements displaced transcriptionally active enhancer sequences close to BCL11B without producing fusion genes. All translocations resulted in the activation of BCL11B, a regulator of T-cell differentiation associated with transcriptional corepressor complexes in mammalian cells. The expression of BCL11B behaved as a disease biomarker that was present at diagnosis, but not in remission. Deregulation of BCL11B co-occurred with variants at FLT3 and at epigenetic modulators, most frequently the DNMT3A, TET2, and/or WT1 genes. Transcriptome analysis identified a specific expression signature, with significant downregulation of BCL11B targets, and clearly separating BCL11B AL from AML, T-ALL, and ETP-ALL. Remarkably, an ex vivo drug-sensitivity profile identified a panel of compounds with effective antileukemic activity.
© 2021 by The American Society of Hematology.
Figures



Comment in
-
BCL11B, the Cerberus of human leukemia.Blood. 2021 Sep 2;138(9):741-743. doi: 10.1182/blood.2021011856. Blood. 2021. PMID: 34473234 No abstract available.
References
-
- Swerdlow SH, Campo E, Harris NL, et al. . WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed., revised. Lyon, France: World Health Organization; 2017.
-
- Patrick K, Wade R, Goulden N, et al. . Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003. Br J Haematol. 2014; 166(3):421-424. - PubMed
-
- Conter V, Valsecchi MG, Buldini B, et al. . Early T-cell precursor acute lymphoblastic leukaemia in children treated in AIEOP centres with AIEOP-BFM protocols: a retrospective analysis. Lancet Haematol. 2016; 3(2):e80-e86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

